Figures & data
Table 1. Bifidobacterium kashiwanohense associated strain listCitation26,Citation27,Citation30–37.
Figure 1. General genome features of B. kashiwanohense.
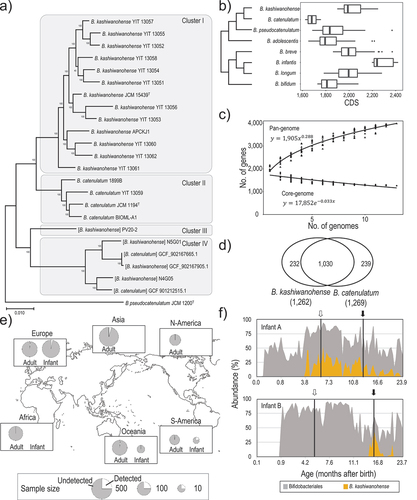
Figure 2. B. kashiwanohense glycobiome and associated growth profiles.
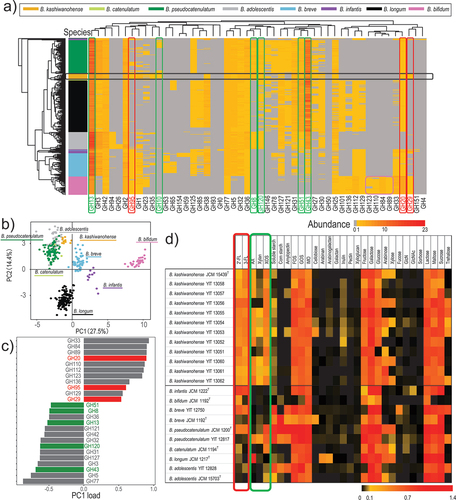
Figure 3. Utilization of xylan-associated carbohydrates by B. kashiwanohense.
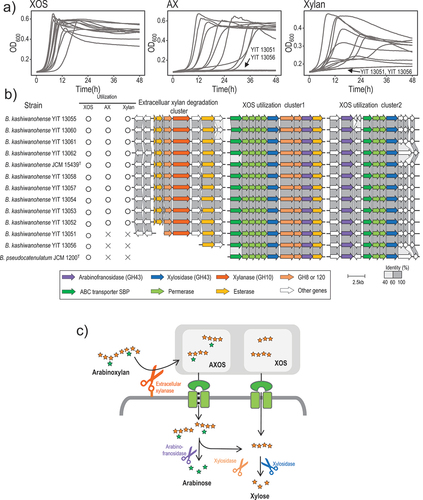
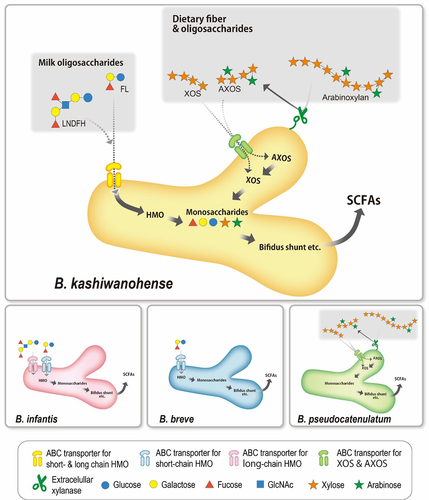
Figure 4. HMO utilization by B. kashiwanohense.
Figure 5. B. kashiwanohense strains utilize both milk- and plant-derived carbohydrates.
Supplemental Material
Download MS Excel (148.4 KB)Supplemental Material
Download PDF (337.4 KB)Data availability statement
The bifidobacterial genome sequences have been deposited in the NCBI Sequence Read Archive under BioProject Accession Code PRJNA883016 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA883016)
