Figures & data
Table 1. Different approaches to the microbiome ML limitations discussed in the introduction.
Table 2. Table of datasets.
Figure 1. iMic’s and gMic’s architectures and AUCs.
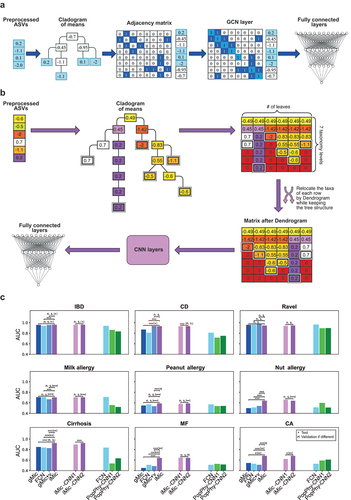
Table 3. Sequential datasets details.
Table 4. 10 CVs mean performances with standard deviation on external test sets; the std is the std among CV folds.
Table 5. Features can be added to iMic’s learning. Average AUCs of iMic-CNN2 with and without non-microbial features as well as average results of naive models with non-microbial features. The results are the average AUCs on an external test with 10 CVs their standard deviations (stds).
Figure 2. iMic copes with the ML challenges above better than other methods.
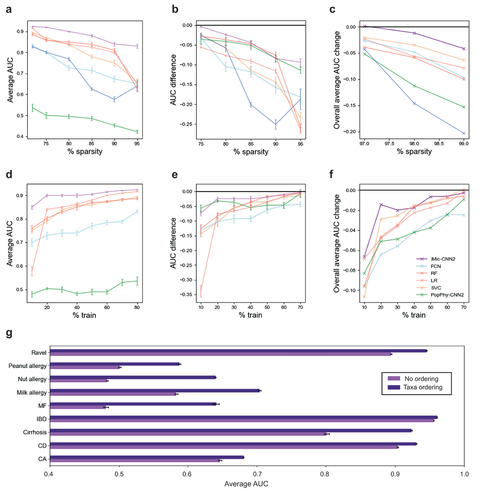
Figure 3. Interpretation of iMic’s results.
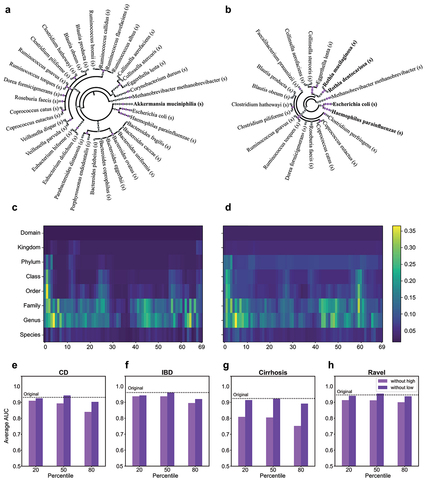
Figure 4. 3D learning.
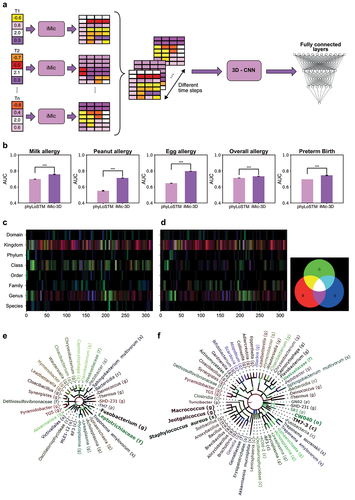
Table 6. Notations.
Table
Table
Table
Supplemental Material
Download MS Word (3.6 MB)Data availability statement
All datasets are available at https://github.com/oshritshtossel/iMic/tree/master/Raw_data.
