Figures & data
Table 1. Participants characteristics.
Figure 1. Dietary intakes and gut microbial composition differed by IBS subtypes.
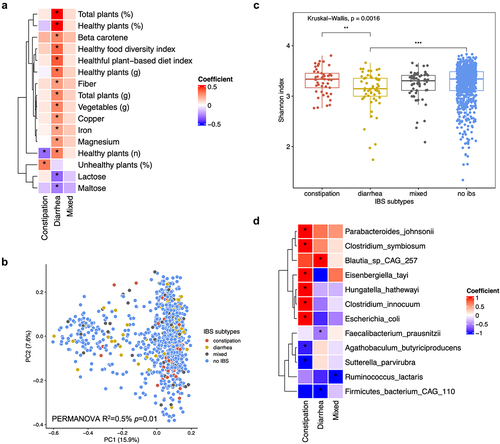
Figure 2. Random forest model classifying IBS subtypes according to gut microbial taxa, functional pathways, and host factors.
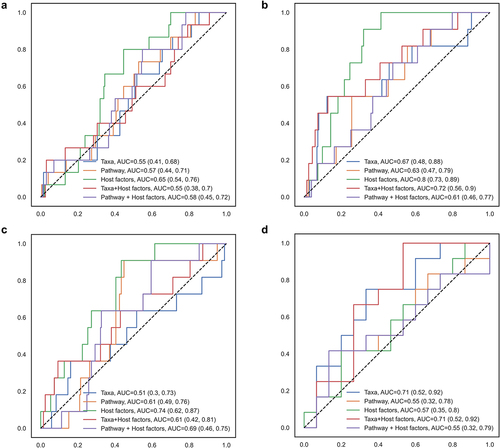
Figure 3. Taxonomic-level results were comparable with prior studies.
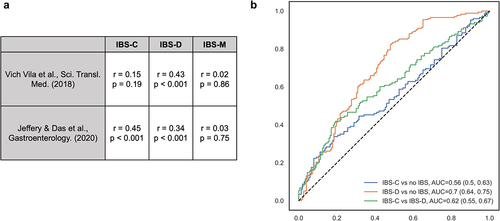
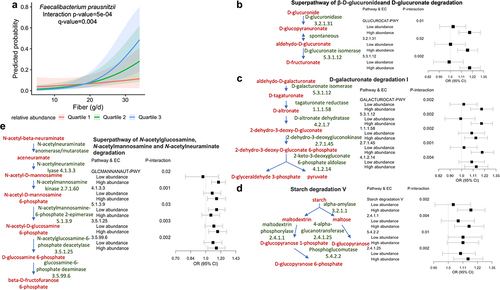
Figure 4. The association between dietary fiber and IBS-D varied by the relative abundance of Faecalibacterium prausnitzii.
Supplemental Material
Download PDF (4 MB)Supplemental Material
Download MS Excel (316.4 KB)Data availability statement
Gut metagenome sequencing data can be accessed from the European Nucleotide Archive (accession number: PRJEB39223). Other data, analytical methods, and study materials can be made available to collaborators upon reasonable request from https://twinsuk.ac.uk/resources-for-researchers/access-our-data/.
