Figures & data
Figure 1. Summary of study design (a) and sample groups (b). All participant stool samples were PCR-screened for DEC pathotype genes and positive isolates were whole-genome sequenced. A random subset of age and sex-matched participant stool samples were selected for 16S rRNA gene amplicon gut microbiome sequencing, and a further subset were selected for shotgun metagenome sequencing so that there would be an approximately equal number of shotgun metagenomes for diarrheal case and control samples and DEC-positive and negative samples. Symbols in panel b indicate diarrhea case (+) or control (-) status and the presence (+) or absence (-) of DEC in participant stools. All samples with a DEC isolate genome are from infections because by-definition there were no DEC isolates detected in the uninfected stool samples. See text and Supplementary Table 2 for additional sample selection and filtering criteria.
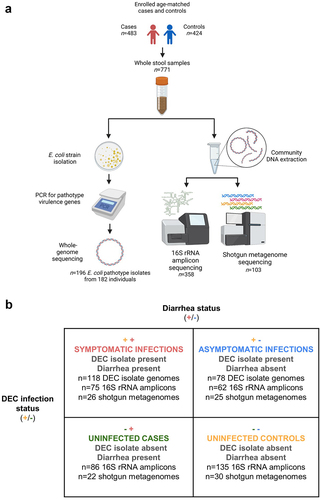
Figure 2. DEC isolate metagenome abundances. Dashed lines indicate mean abundance for case versus control sample groups. Data are also shown as an inset box plot.
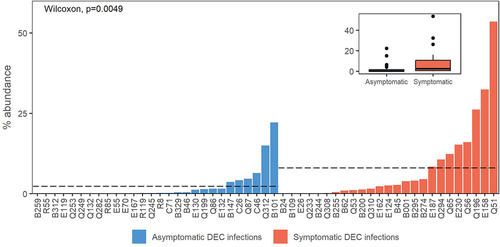
Figure 3. Comparison of the numbers of virulence genes from the virulence factor database (VFDB) in symptomatic versus asymptomatic DEC infections and in uninfected cases versus controls. Data are shown for virulence genes in the VFDB annotated as any taxa (a) and as E. coli (b).
Figure 4. Mean relative abundances of virulence genes that were significantly differentially abundant by DEC infection and diarrhea case/control status (4-way Kruskal-Wallis adjusted p-values < 0.05).

Figure 5. Comparisons of coverage-based estimates of within-sample alpha diversity for symptomatic versus asymptomatic DEC infections and for uninfected cases versus controls. Shannon, Simpson, and observed alpha diversity were calculated using 16S rRNA gene amplicon data (a-c); Nonpareil alpha diversity was calculated using shotgun metagenome data (d).
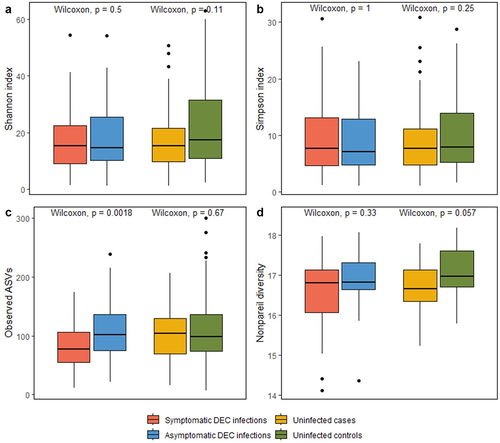
Figure 6. Mean relative abundances of 16S rRNA gene amplicon family-level bacterial taxa that were significantly associated with diarrhea and DEC infection status (corncob and/or LEfSe analyses; adjusted p < 0.05, LEfSe LDA threshold < 3). Values are square root transformed to improve visualization of taxa with low relative abundances.
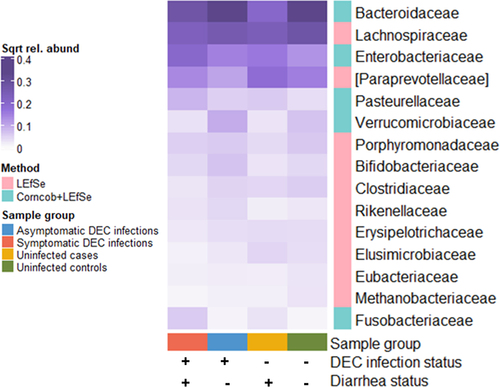
Figure 7. Mean relative abundances of genes with secondary KEGG annotations that were significantly differentially abundant by DEC infection and diarrhea status (4-way Kruskal-Wallis adjusted p-values < 0.05).
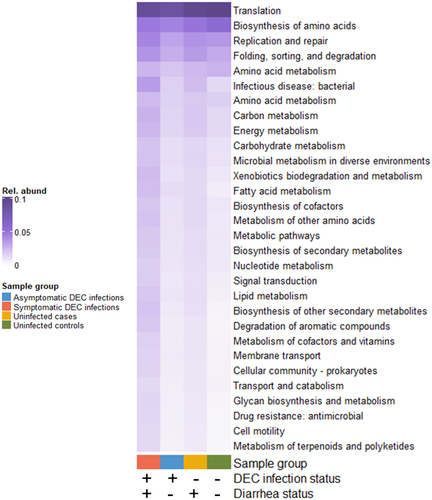
Figure 8. Core genome phylogenetic tree of DEC strains from symptomatic and asymptomatic individuals; there was no phylogenetic signal for case/control status (δ = 0.59, p = 0.92).
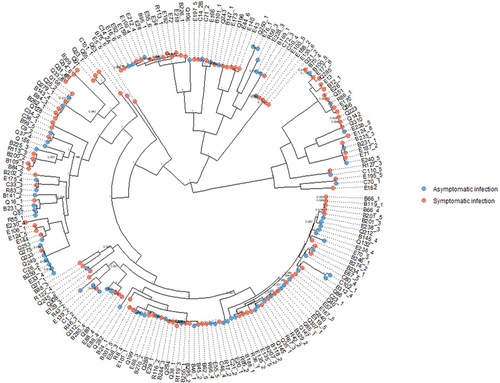
Table 1. Results summary for analyses comparing pathogen abundance, gut microbiome characteristics, and pathogen characteristics for symptomatic versus asymptomatic DEC infections.
Gut Microbes supplement 2nd resubmission clean.docx
Download MS Word (4.2 MB)Data availability statement
The data that support the findings of this study are openly available under NCBI BioProject PRJNA486009. R code is available on Github: https://github.com/kjojess/EcoZUR-symptomatic-asymptomatic-manuscript.
