Figures & data
Figure 1. Derivation of organoid lines from a cohort of healthy individuals.
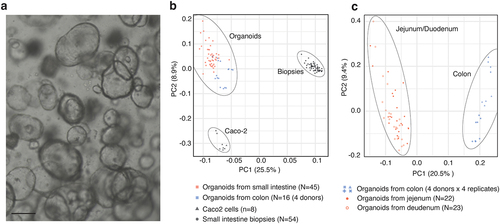
Figure 2. Characterization of impact of media composition on organoid cultures.

Figure 3. Development and characterization of a 2D model for primary intestinal epithelial cells.
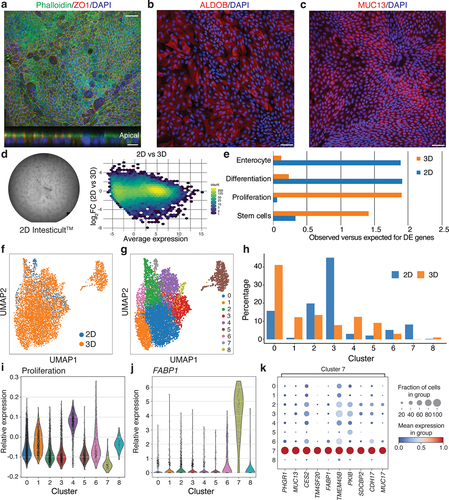
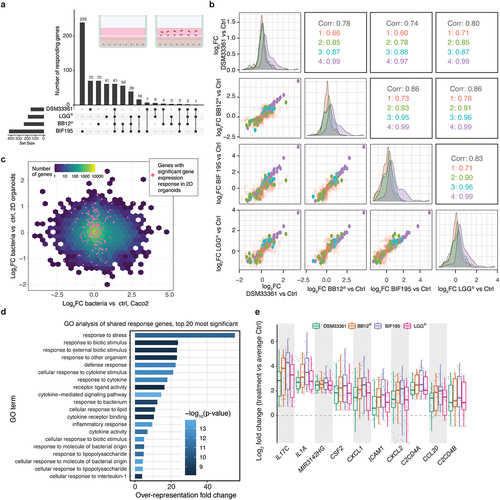
Figure 4. Response to coculture with different microbes.
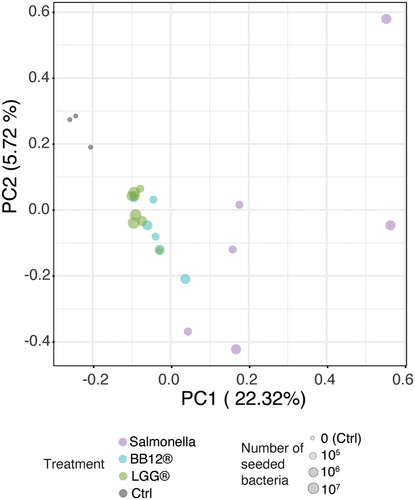
Figure 5. Screening bacteria using 2D model for primary intestinal epithelial cells.
Supplemental Material
Download Zip (11 MB)Data availability statement
The expression data from CaCO2 cells stimulated with bacterial isolates are deposited at the NCBI Gene Expression Omnibus (GEO) under accession number GSE231605. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE231605
