Figures & data
Figure 1. Experimental design.
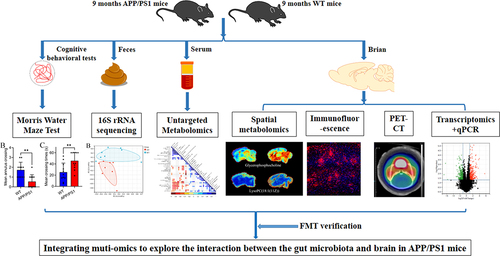
Figure 2. Cognitive level and neuropathology in APP/PS1 and WT mice.
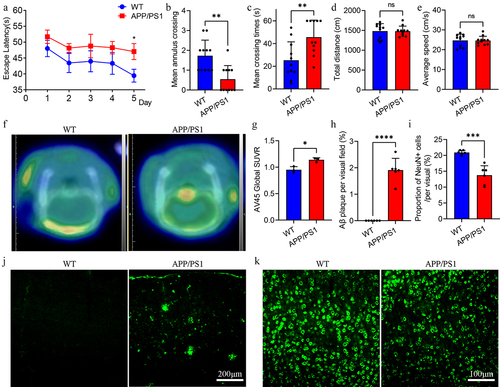
Figure 3. The difference of gut microbiota between APP/PS1 and WT mice.
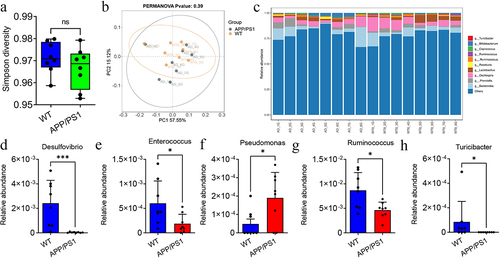
Figure 4. The difference of metabolites in serum and brain of APP/PS1 and WT mice.
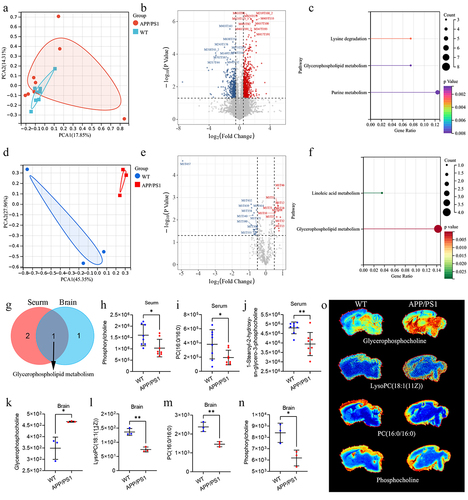
Figure 5. Difference analysis of neuroinflammation and signaling pathway in APP/PS1 and WT mice.
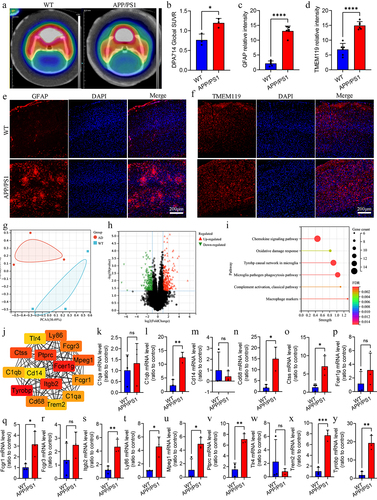
Figure 6. Correlation analysis of different gut microbiota, metabolites and hub differently expressed genes.
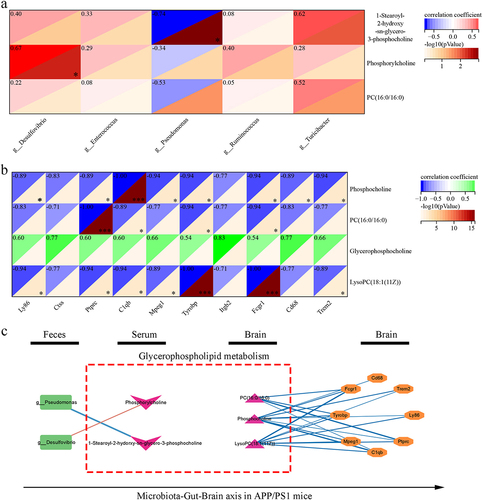
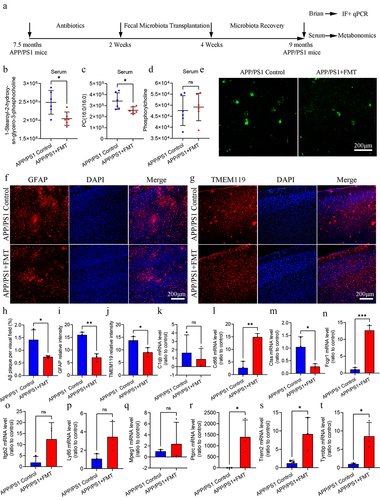
Figure 7. FMT intervention regulated glycerophospholipid metabolism and ameliorated Aβ pathology and neuroinflammation in APP/PS1 mice.