Figures & data
Figure 1. Application of a mouse model of IBD to test the effect of gut inflammation on the evolution of NC101.
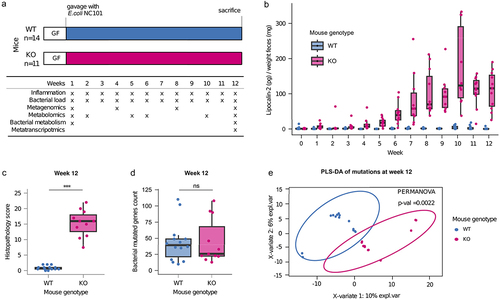
Figure 2. Parallel evolution of E. coli in inflamed mice revealed a disease-specific genetic signature.
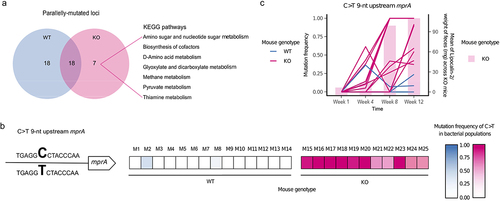
Figure 3. Phenotypic characterization of evolved E. coli and the inflammatory environment in which they were selected.
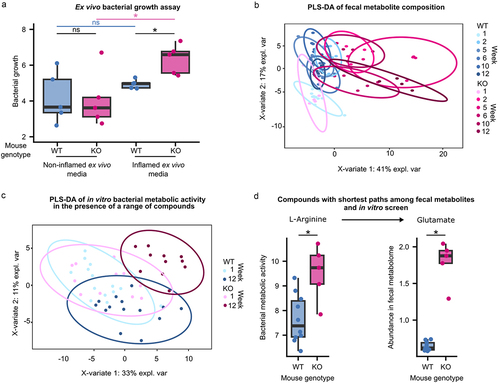
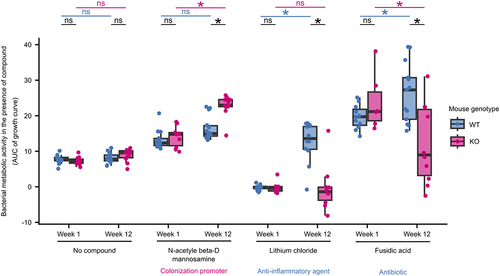
Figure 4. E. coli adapted to the inflamed gut show phenotypes with clinical relevance.
Supplementary_tables_GutMicrobes_rev1.xlsx
Download MS Excel (2.6 MB)Supplementary_figures_GutMicrobes_rev1 clean.docx
Download MS Word (2.5 MB)Data availability statement
Escherichia coli NC101 reference genome is available under Bioproject number PRJNA596436. Shotgun metagenomic sequencing of the inocula and evolved populations and metatranscriptomic of the populations at week 12 are available with Bioproject number PRJNA1012288.
