Figures & data
Figure 1. Phylogeny and competition factors of ETEC strains.
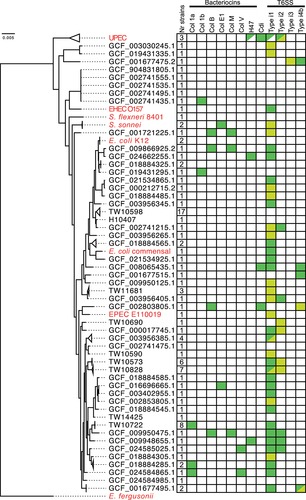
Figure 2. Conservation and selective pressure of type i1 T6SS components in ETEC strains with potentially active systems.
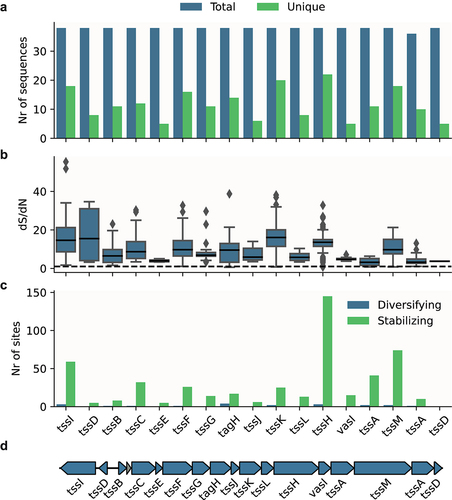
Figure 3. Representative domain composition of putative T6SS effectors identified in ETEC strains with potentially active T6SS (N = 47).
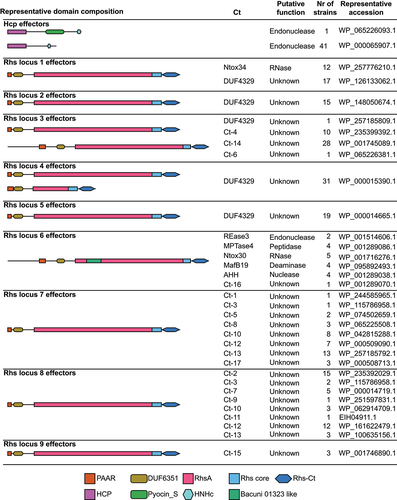
Figure 4. ETEC TW10722 uses Colicin Ia to outcompete MG1655 in vitro.
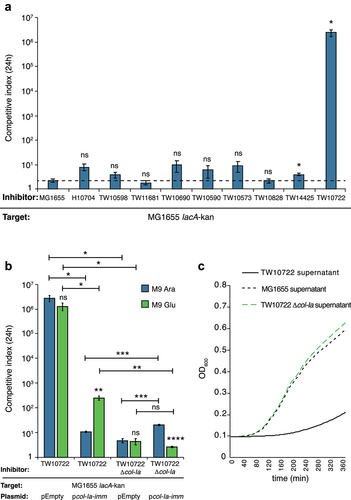
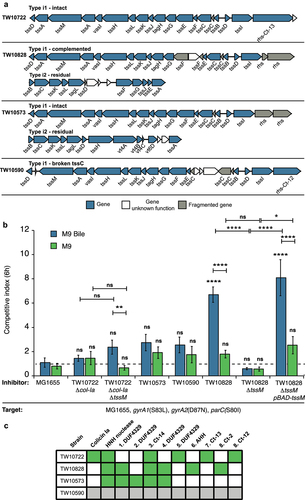
Figure 5. ETEC TW10828 uses T6SS to outcompete MG1655 in vitro.
Table_S11.xlsx
Download MS Excel (19 KB)Table S7 revised.xlsx
Download MS Excel (98.5 KB)Table_S12.xlsx
Download MS Excel (51.2 KB)Table_S2_revised_JK.xlsx
Download MS Excel (29.7 KB)Table S6 revised.xlsx
Download MS Excel (370.3 KB)Table S5 revised.xlsx
Download MS Excel (134.9 KB)Supplemental_material_revised 231201 clean.docx
Download MS Word (1.3 MB)Data availability statement
All raw data for the manuscript are freely available either as supplementary excel files (Tables S5–8) or at the NCBI Reference Sequence Database (GCF_032367395.1, GCF_032365155.1, GCF_032366635.1, GCF_032368225.1).
