Figures & data
Figure 1. Schematic overview of the key experimental steps carried out to screen for novel phage-host pairs targeting Bacteroidales order bacteria.
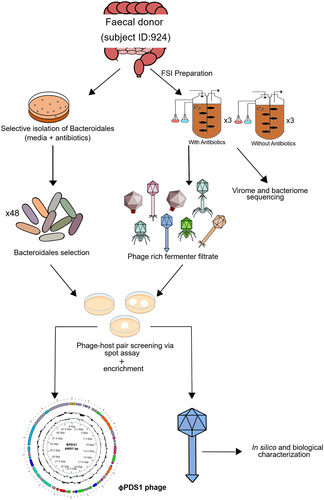
Figure 2. Circular genome map of the novel phage φPDS1 with a genome size of 44,691 bp. The innermost ring represents the GC skew (blue for the positive strand and green for the negative strand) while the central ring (black) displays the GC content. The outermost circle illustrates the coding genes (CDS) with HHpred predicted function as labels. The coloration of the CDS corresponds to their general functions as indicated in the legend. Genes whose function could not be determined are colored gray and remain unlabeled.
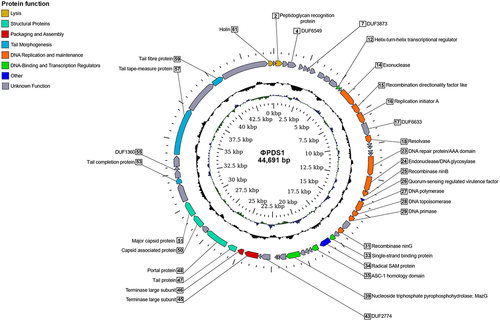
Figure 3. Phylogenomic genome BLAST distance phylogeny (GBDP) tree representing φPDS1, 78 members of the new candidate Paboviridae family, and using 19 phages from the Drexlerviridae family as outgroup. The tree was generated using the VICTOR tool with the D0 formula. The numbers above the branches indicate GBDP pseudo-bootstrap support values from 100 replicates. The branch lengths are scaled in terms of the respective distance formula used. Family, genus, species are tentatively grouped by their phylogenetic relationships when the same shape and color coincide. The GC content is represented in blue; the darker the color, the higher the content. Accession numbers are provided in brackets.
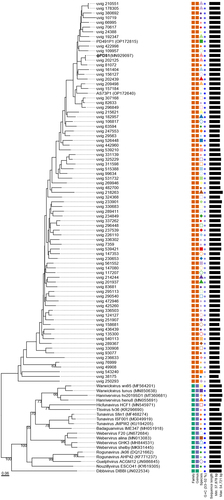
Figure 4. Whole genome comparisons of φPDS1 against members of the different genus clusters within the candidate Paboviridae family identified by VIRIDIC, along with one outgroup member from the Drexlerviridae family (KY619305; Nouzillyvirus ESCO41). The figure shows the percentage of identity of different proteins compared to the closest phylogenetic phage genome. The comparison was performed using Diamond. The color between two genes represents the percentage of identity between both at the protein level. The color of the gene is based on the annotation given by Pharokka. The left-side tree was inferred using the VICTOR tool and the figure was created using the R packages gggenomes and ggtree.
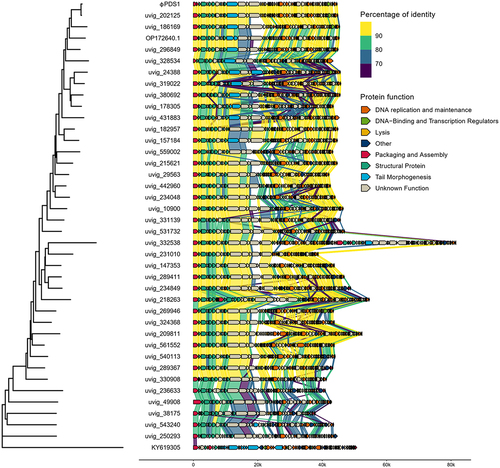
Figure 5. Abundance of φPDS1 in the fermenter vessel with and without selective conditions showing that the selective conditions greatly aided φPDS1 propagation. (a) Relative abundance of φPDS1 with and without antibiotics obtained from the total viral reads sequenced. (b) Absolute quantification of φPDS1 via qPCR targeting a segment of the phage DNA polymerase. Titre determined in copies/ml using the standard curve method. Error bars represent standard deviation (n = 3). * indicates statistically significant differences between vessels, with a p-value <0.05 after performing an unpaired t-test or the Mann – Whitney U test, depending on the data distribution (normal or non-normal, respectively).
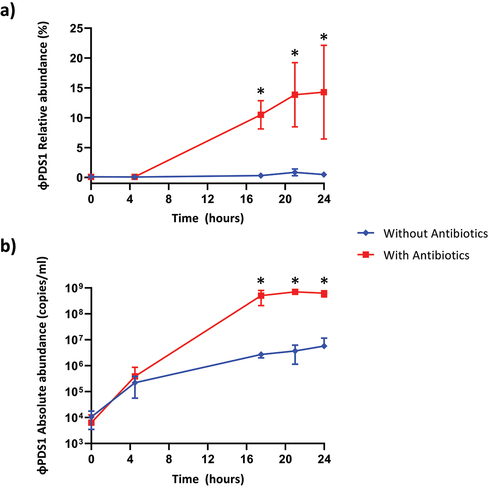
Figure 6. Characterisation and visualization of φPDS1 and formed spots and plaques. (a) Transmission electron micrograph of φPDS1 generated from the enriched lysate, stained with uranyl acetate showed that φPDS1 has a siphovirus morphology. The capsid diameter is approximately 53 ± 2.0 nm, and the tail length is 150 ± 10.0 nm. (b) Spot morphology with incomplete clearing. (c) Visualization of φPDS1 plaques from a spot assay at different titers using a stereoscopic microscope.
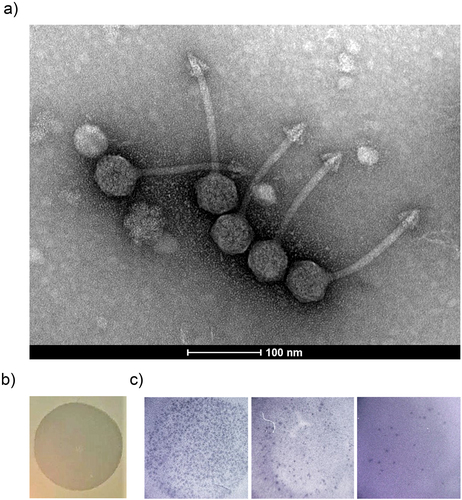
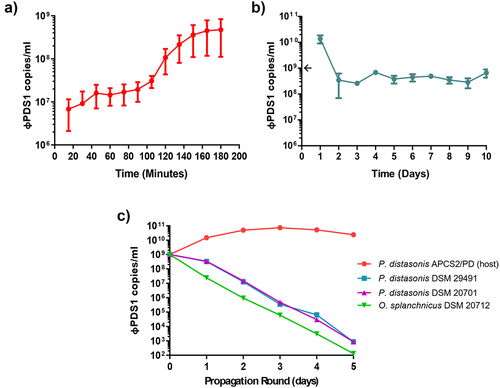
Figure 7. Biological characterization of φPDS1. (a) φPDS1 one-step growth curve. Sampling was performed every 15 minutes over 3 hours. The latent period was 90 minutes, and the burst size was ~ 23 copies per infected cell. (b) The titer of φPDS1 following continuous co-culture on the host over 10 days showing the persistence of the phage with its host. (c) Liquid propagation of φPDS1 on two commercial P. distasonis strains (DSM 29491 and DSM 20701) and O. splanchnicus (DSM 20712) was examined over five days with host P. distasonis APCS2/PD used as a control. Titres were determined in copies/ml via qPCR for all the experiments. Arrows on the y-axis indicate phage titer, after dilution, on initiation of the experiment. Error bars indicate standard deviation (n = 3).
Supplemental Material
Download MS Excel (11.4 KB)Supplemental Material
Download MS Excel (16.2 KB)TableS3.xlsx
Download MS Excel (10.1 KB)TableS1.xlsx
Download MS Excel (11.3 KB)TableS7.xlsx
Download MS Excel (45.4 KB)TableS2.xlsx
Download MS Excel (14.1 KB)TableS4.xlsx
Download MS Excel (781.3 KB)SupplementaryMaterial_PDS1_submission_clean.docx
Download MS Word (14.2 MB)Data availability statement
All data generated or analyzed during this study are included in this report and in the supplementary material. The genome of φPDS1 is deposited into GenBank under accession MN929097 (assembled and annotated genome). The genome of the bacterial host of φPDS1, Parabacteroides distasonis APCS2/PD, is deposited under the following accession codes: BioProject PRJNA556872, GenBank CP042285 (assembled and annotated genome) and associated plasmid pPDS2–1, GenBank CP042284.
