Figures & data
Figure 1. Human data.
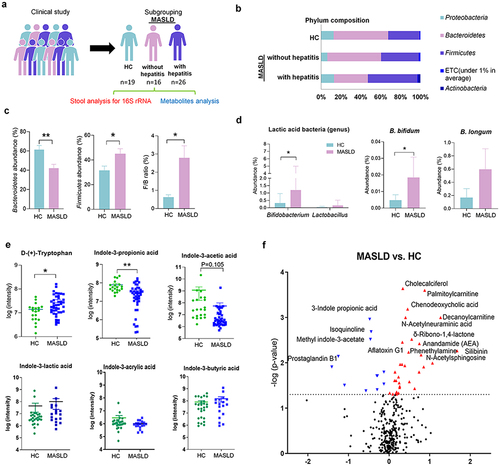
Figure 2. IPA and IAA ameliorate WD-induced hepatic steatosis.
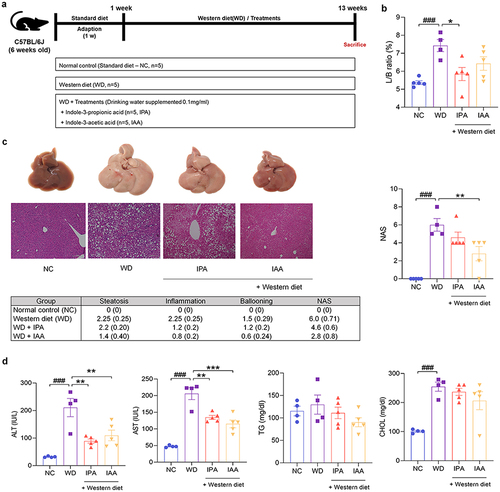
Figure 3. Administration of IPA and IAA alleviates WD-induced hepatic steatosis and inflammation in mice.
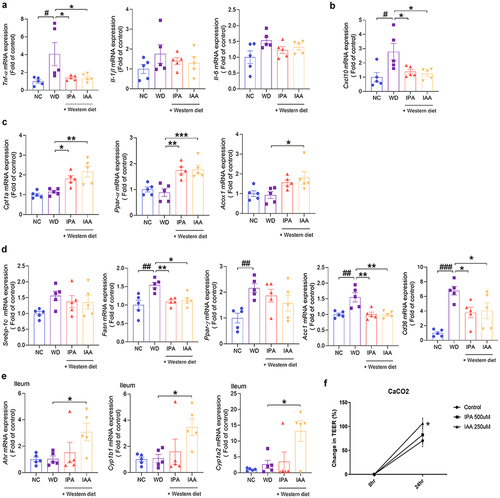
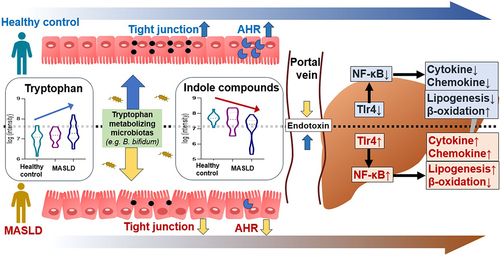
Figure 4. Administration of IPA and IAA suppresses the increase in endotoxin and the inflammatory response caused by WD.
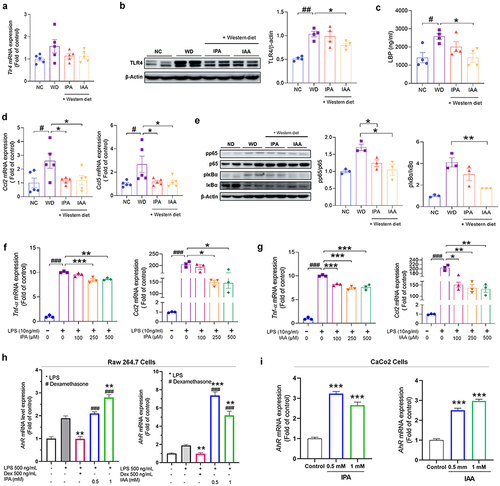
Figure 5. IPA and IAA treatment altered transcriptomic in the liver.
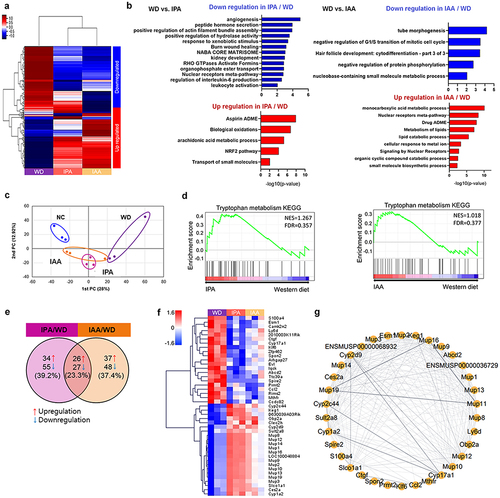
Figure 6. Modulating microbial taxonomic abundance of IPA and IAA on WD-induced changes.
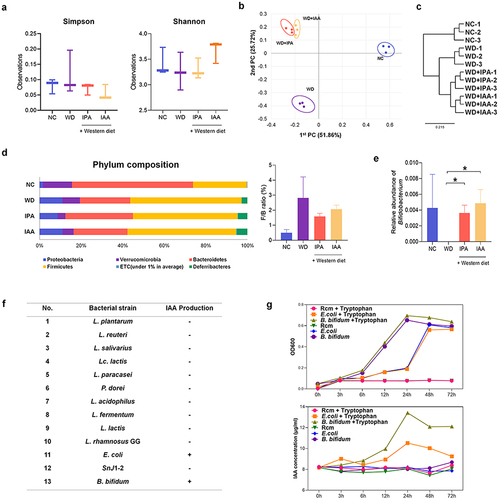
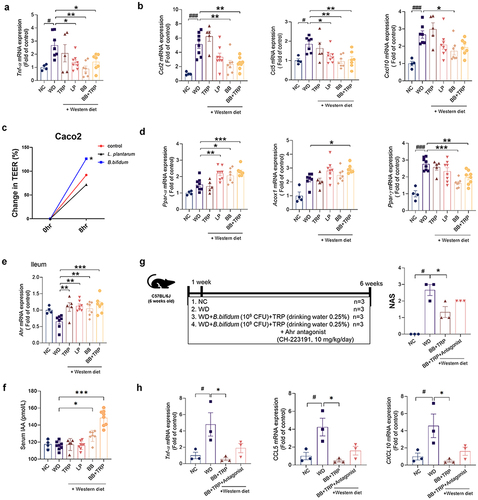
Figure 7. B. bifidum ameliorates WD-induced hepatic steatosis.
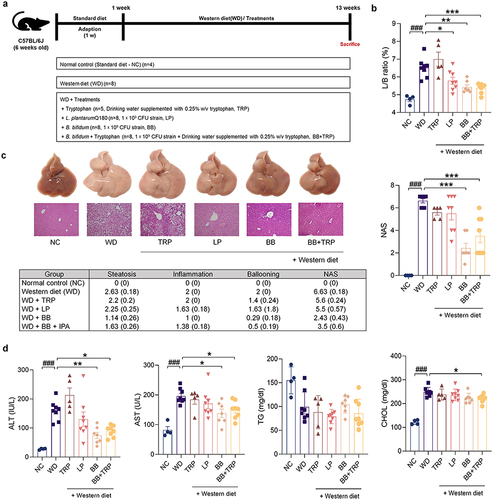
Figure 8. Administration of B. bifidum alleviates WD-induced hepatic steatosis and inflammation in mice.