Figures & data
Figure 1. Dysbiosis of gut microbiota in patients with preeclampsia (PE). (a) Comparison of the α diversity of the gut microbiota between the preeclampsia (PE) and normal pregnant women (NP) groups. (b) Comparison of the β diversity of gut microbiota between the PE and NP groups. (c) Venn diagram of operational taxonomic units (OTUs) in the PE and NP groups. (d) Differences in the structure and Amplicon sequence variant (ASV)/OTU ratio of gut microbiota at the phylum level between the PE and NP groups. (e) Differences in the structure and ASV/OTU ratio of gut microbiota at the genus level between the PE and NP groups. (f) Bar plot of the linear discriminant analysis (LDA) value distribution of significantly different species, showing the significantly enriched species and their importance degree in the two groups. Data are presented as the median and quartile in panel A, and statistical analysis was performed using the t-test. *p < .05, **p < .01, ***p < .001. NP group: n = 29; PE group: n = 38.
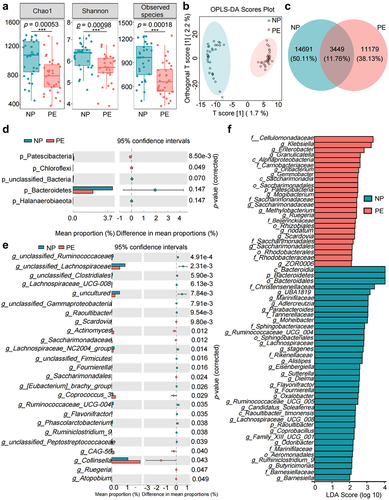
Figure 2. Changes in Trimethylamine N-oxide (TMAO) and its precursors in patients with PE, and source analysis of the microbiota. (a) Differences in TMAO and its precursor levels between the PE and NP groups by orthogonal partial least squares discriminant analysis (OPLS-DA). (b) Heat map of serum TMAO and its precursor expression levels in the PE and NP groups. (c) Quantitative analysis of serum TMAO and its precursors in the NP and PE groups. (d) Correlation analysis between serum TMAO and its precursors and key clinical indicators of PE. (e) The ability of TMAO and its precursors to diagnose PE was analyzed by receiver operating characteristic (ROC) curve. (f) Difference in the abundance of TMAO-producing bacteria between the NP and PE groups. (g) The relative abundance of klebsiella, Gemmobacter, and Ruegeria of the PE and NP groups. (h) Correlation analysis between TMAO-producing bacteria and PE clinical indicators. Data are presented as the mean ± SD in C and F plots, and statistical analysis was performed using the t-test. *p < .05, **p < .01, ***p < .001. NP group: n = 29; PE group: n = 38. SBP: systolic blood pressure, DBP: diastolic blood pressure, GW: gestational weeks, FBW: fetal birth weight, WBC: white blood cell, NEU: neutrophils, TBA: total bile acid, HC: head circumference, BPD: biparietal diameter, AC: abdominal circumference, FL: femoral length, U/C: Urea/creatinine, AST/ALT: aspartate aminotransferase/alanine aminotransferase, TT: thrombin time.
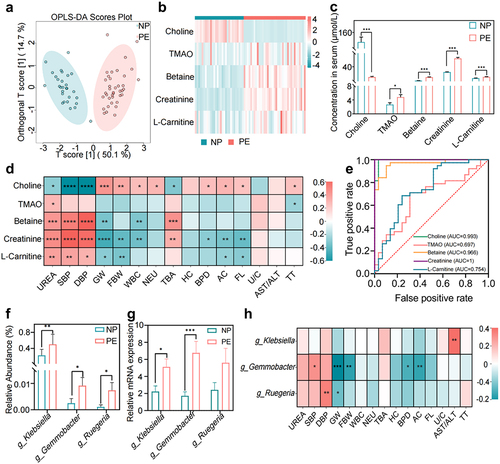
Figure 3. TMAO inhibitor 3,3-dimethyl-1-butanol (DMB) reversed the PE phenotype in mice that received fecal microbiota transplantation from patients with PE. (a) PE-fecal microbiota transplantation (FMT) animal experiment protocol. (b) Dynamic changes and comparison of the systolic blood pressure (SBP) in each group of mice (n = 6). (c) Dynamic changes and comparison of the diastolic blood pressure (DBP) among groups (n = 6). (d) Twenty-four-hour urinary protein content before FMT, on day 42 of FMT (before pregnancy), and on day 59 of FMT (day 17 of pregnancy). (e) Whole body photograph of embryo and placenta. (f) Weight of fetal mice. (g) Placental efficiency was assessed by the ratio of embryo to placental weight. (h) Representative images of sagittal tissue sections in hematoxylin-eosin (h&e) stained placental tissue. Regions are marked and represented by maze and junctional zones. (i) Representative H&E staining images of the placental maze area of mice in each group. Data are presented as the mean ± SEM. Significant differences based on one-way ANOVA and Tukey’s multiple tests: *p < .05, **p < .01, ***p < .001, control group compared to the FMT group; ###p < .001, ##p < .01, #p < .05, FMT+DMB group compared to the FMT group.
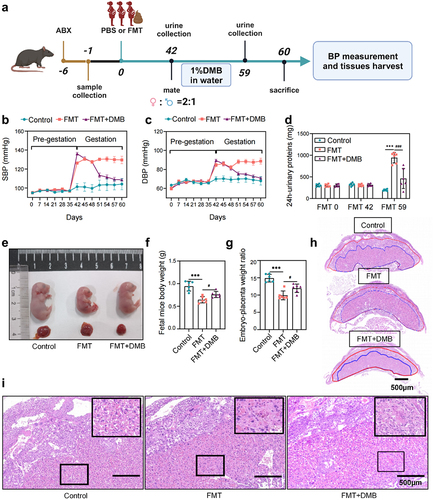
Figure 4. Targeting TMAO significantly attenuated oxidative stress and inflammatory injury in PE-FMT recipient mice. (a) Activity levels of placental oxidative stress injury markers in each group of mice after PE fecal microbiota transplantation. (b) Comparison of serum inflammatory factors (IL-6, TNF-α, IL-1β) levels. (c-d) Western blot was used to detect the expression levels of Nrf2-Keap1 signaling pathway-related molecules in the placental tissues of mice in each group. (e-f) Western blot was used to detect the expression of MAPK family kinases (P38, p-P38, ERK, p-ERK, JNK, p-JNK) signaling pathway-related molecules in the placental tissues of each group. (g) Immunohistochemistry was used to detect the expression levels of MAPK family kinases (p-P38, p-ERK, p-JNK) and Nrf2-Keap1 signaling pathway-related molecules in placental tissues of each group. Data are presented as the mean ± SEM. Significant differences based on one-way ANOVA and Tukey’s multiple tests: *p < .05, **p < .01, ***p < .001, control group compared to the FMT group; ###p < .001, ##p < .01, #p < .05, FMT+DMB group compared to the FMT group n = 6.
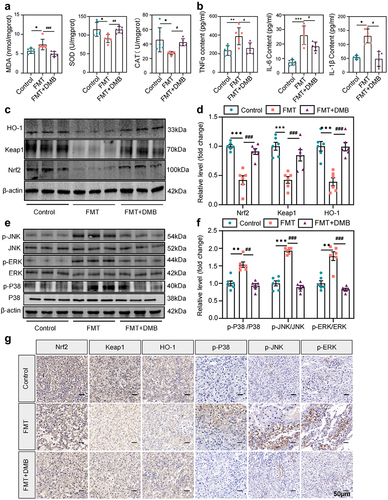
Figure 5. Targeted intervention with TMAO reduces the clinical phenotype of NG-Nitroarginine methyl ester (L-NAME)-induced PE. (a) Animal experimental protocol of L-NAME-induced PE model mice. (b) Dynamic changes and comparison of the SBP in each group during pregnancy (n = 6). (c) Dynamic changes and comparison of the DBP in each group during pregnancy (n = 6). (d) The urine of G10 and G17 mice in each group was collected, and the urinary protein concentration and 24-h urinary protein content were measured. (e) Whole-body photographs of fetal mice and placentas of the three groups. (f) Weight of the fetal rats. (g) The placental efficiency of the three groups was assessed by the ratio of fetal to placental weight. (h) Representative images of placental tissues stained with H&E. Regions are marked and represented by maze and junctional zones. (i) Representative H&E staining images of the placental maze area of each group of mice. Data are presented as the mean ± SEM. Significant differences based on one-way ANOVA and Tukey’s multiple tests: *p < .05, **p < .01, ***p < .001, control group compared to the L-NAME group; ###p < .001, ##p < .01, #p < .05, L-NAME+DMB group compared to the L-NAME group.
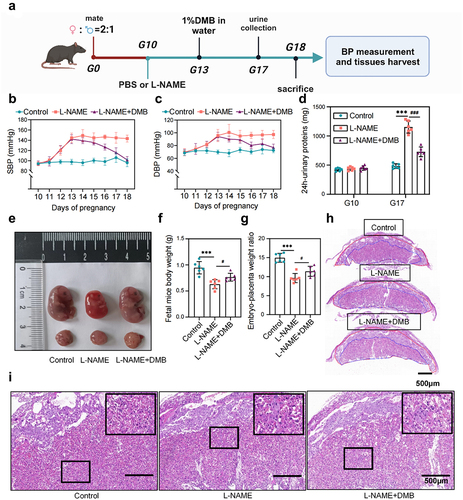
Figure 6. Targeted intervention with TMAO attenuated oxidative stress and inflammation in L-NAME-induced PE mice. (a) Activity levels of placental oxidative stress injury markers in each group of mice. (b) Comparison of serum inflammatory factor (IL-6, TNF-α, IL-1β) levels. (c–d) expression levels of Nrf2-Keap1 signaling pathway-related molecules in placental tissues. (e–f) expression levels of MAPK family kinase (P38, p-P38, ERK, p-ERK, JNK, p-JNK) signaling pathway-related molecules in placental tissues. (g) Immunohistochemistry was used to detect the expression levels of MAPK family kinases (p-P38, p-ERK, p-JNK) and Nrf2-Keap1 signaling pathway-related molecules in the placental tissues of each group. Data are presented as the mean ± SEM (A–F). *p < .05, **p < .01, ***p < .001, control group compared to the L-NAME group; ###p < .001, ##p < .01, #p < .05. The comparison between the L-NAME+DMB group and the L-NAME group was performed by one-way ANOVA and Tukey’s multiple tests; n = 6.
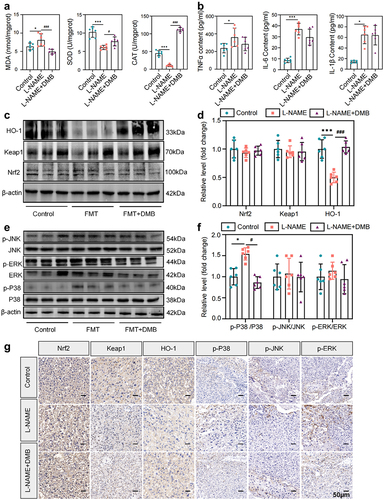
Figure 7. TMAO promotes oxidative stress and inflammatory injury and inhibits HUVEC migration and angiogenesis. (a) Fluorescence imaging was used to detect the intracellular reactive oxygen species (ROS) level in HUVECs after TMAO treatment. (b-c) flow cytometry was used to detect the intracellular ROS level of HUVECs after TMAO treatment. (d-e) expression levels of molecules related to the Nrf2-Keap1 signaling pathway. (f – g) expression levels of molecules associated with MAPK family kinase (P38, p-P38, ERK, p-ERK, JNK, p-JNK) signaling pathways. (h-j) TMAO affects the migration ability of HUVECs. (l-k) TMAO affects the angiogenic ability of HUVECs. Data are presented as the mean ± SEM. Differences between the mean values of the normally distributed data were analyzed using the wilcoxon rank sum test. *p < .05, **p < .01, ***p < .001, control group compared to the TMAO group; ###P < .001, ##P < .01, #P < .05, respectively, the comparison between the NAC, TMAO+NAC, and TMAO groups; n = 3.
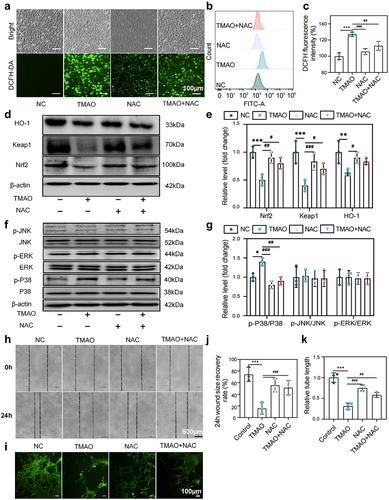
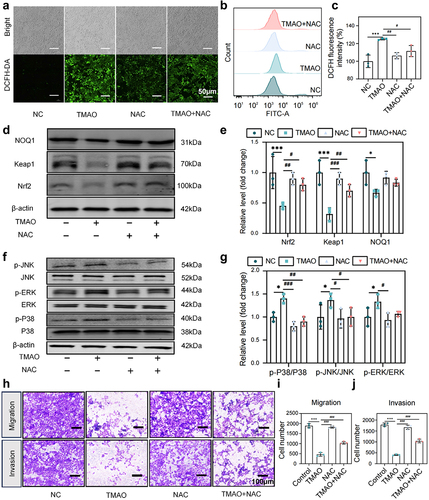
Figure 8. TMAO inhibits the migration and invasion of HTR-8/SVneo cells by activating the MAPK/Nrf2-Keap1 signaling pathway. (a) Fluorescence imaging was used to detect the intracellular ROS level in HUVECs after TMAO treatment. (b-c) flow cytometry was used to detect the intracellular ROS level of HUVEC after TMAO treatment. (d-e) expression levels of molecules related to the Nrf2-Keap1 signaling pathway. (f-g) expression levels of molecules associated with MAPK family kinase (P38, p-P38, ERK, p-ERK, JNK, p-JNK) signaling pathways. (h) TMAO affected the migration and invasion of HTR-8/SVneo cells. Data are presented as the mean ± SEM. Differences between the mean values of the normally distributed data were analyzed using the wilcoxon rank sum test. *p < .05, **p < .01, ***p < .001, control group compared to the TMAO group;###p < .001, ##p < .01, #p < .05, respectively, comparison between the NAC, TMAO+NAC, and TMAO groups; n = 3.
Supplemental Material
Download Zip (3.2 MB)Data availability statement
The data that support these findings of the study are available upon request from the corresponding authors.