Figures & data
Figure 1. Organization of genes within the cag PAI. Five genes encoding proteins localized to the T4SS OMCCCitation10,Citation11 and three genes encoding putative ATPases localized to the T4SS IMC are indicated. Genes required for Cag T4SS activity that lack homologs in other bacterial species are indicated with diagonal stripes.
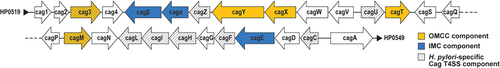
Figure 2. Cag T4SS-mediated delivery of CagA and non-protein substrates into host cells. The Cag T4SS is required for delivery of CagA, LPS metabolites, peptidoglycan and DNA into host cells. Each substrate elicits a cellular response. CagA is phosphorylated by tyrosine kinases (such as Src), and phosphorylated CagA can cause disruptions to a variety of signaling pathways. LPS metabolites and peptidoglycan elicit NF-κB activation, leading to IL-8 production. DNA translocation causes TLR9 activation. Created with BioRender.com.
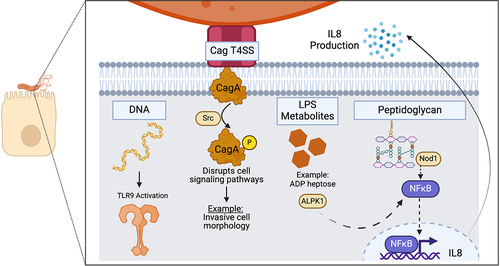
Figure 3. Structural organization of the Cag T4SS OMCC. The 14-fold-symmetric OMC and 17-fold-symmetric PR are illustrated.Citation10,Citation11 (a, b, c) conserved Cag T4SS components. Purple = CagY, yellow = CagX, pink = CagT. (d, e, f) H. pylori-specific T4SS components. Blue = Cag3, green = CagM.
Table 1. cag PAI-encoded proteins that contribute to Cag T4SS activity.
