Figures & data
Figure 1. Bile acid 7-dehydroxylation by C. scindens ATCC 35704.(a) CA 7α-dehydroxylation proposed by Funabashi and colleaguesCitation9 (the six-enzyme pathway) projected onto CDCA. (b) bai operon along with other genes known or suspected to be involved in bile acid transformations in C. scindens ATCC 35704 and the function of the encoded proteins. All enzymes were purified except BaiG.
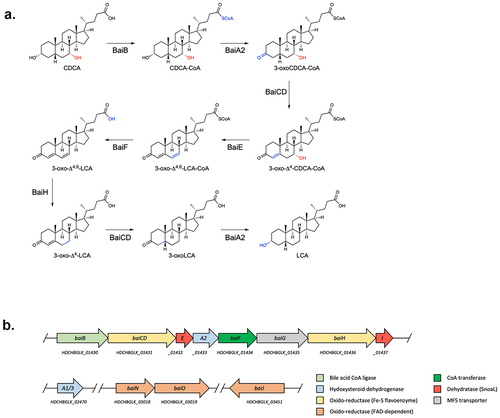
Figure 2. 7α-dehydroxylation of CDCA using different enzyme combinations in vitro.
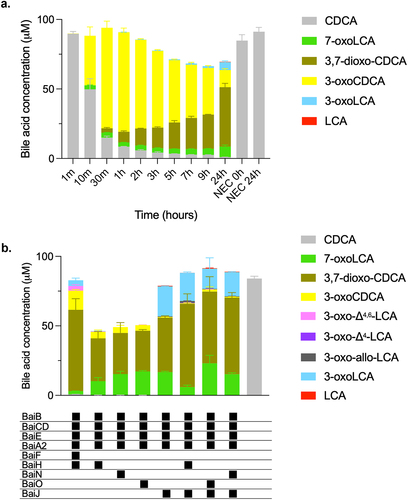
Figure 3. Conversion of CDCA over time with five-enzyme set (BaiB, BaiCD, BaiE, BaiA2, BaiJ).
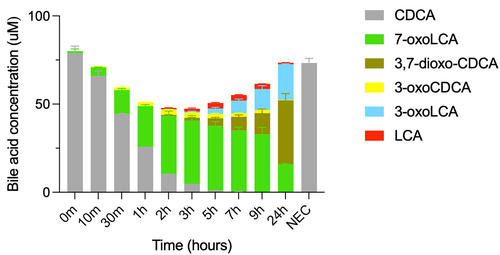
Figure 4. Conversion of CDCA by E. coli expressing bai genes.
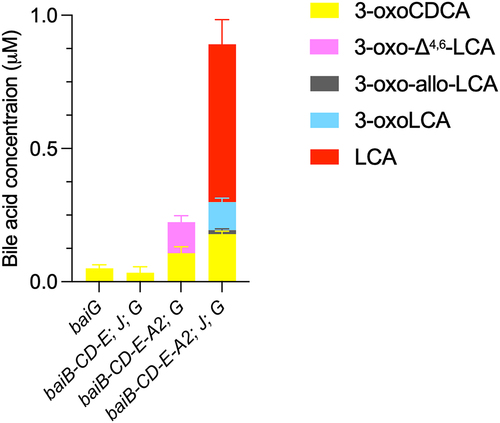
Figure 5. Conversion of CDCA during sequential addition of five-enzyme set enzymes.
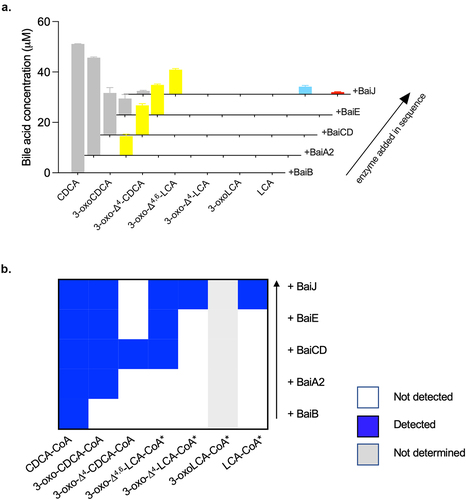
Figure 6. First reductive step in 7α-dehydroxylation pathway.
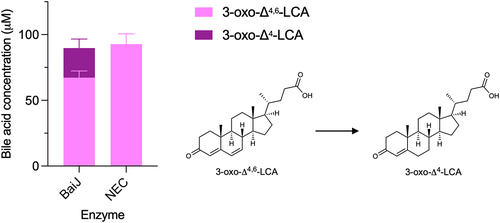
Figure 7. Second reductive step in 7α-dehydroxylation pathway.
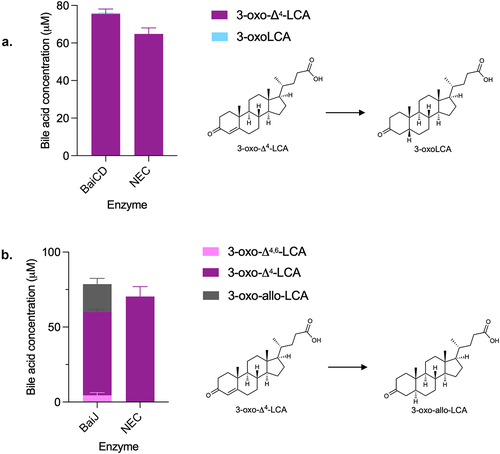
Figure 8. Second reductive step with CoA-ligated bile acids.
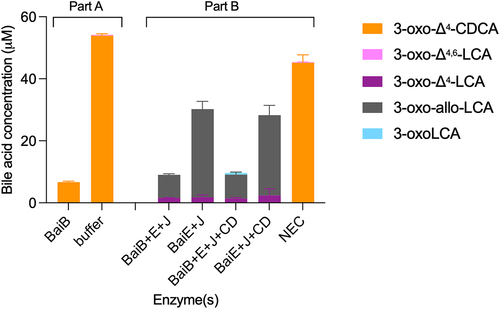
Figure 9. BaiJ from C. scindens strain VPI 12,708 cannot initiate reductive part of 7α-dehydroxylation pathway.
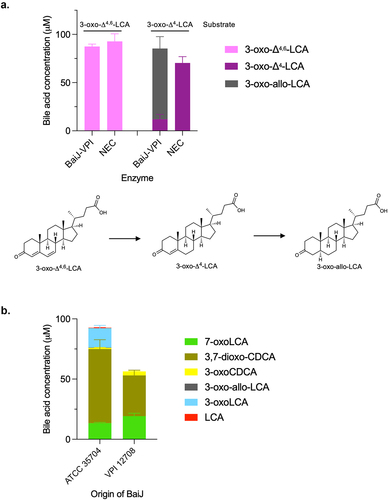
Figure 10. Third reductive step in 7α-dehydroxylation pathway.
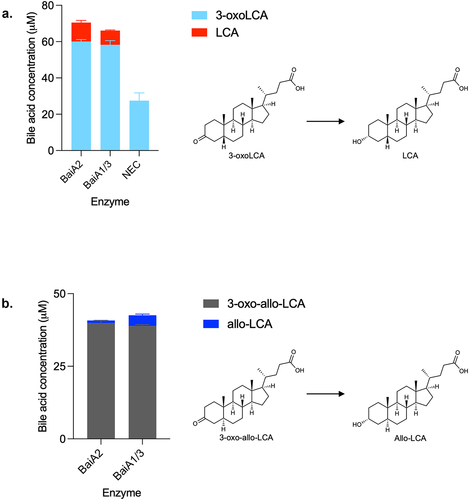
Figure 11. Model of CDCA 7α-dehydroxylation in C. scindens ATCC 35704.
SI_all_revised_final clean.docx
Download MS Word (6.7 MB)Data availability statement
The data used in this manuscript are available at DOI 10.5281/zenodo.8263047
