Figures & data
Figure 1. The validation of the mNFE method on a simulation dataset.
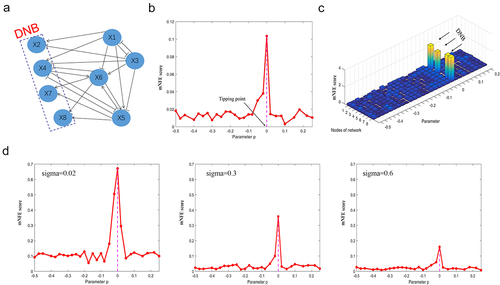
Figure 2. The identification of critical states for seroconversion and T1D based on mNFE at the overall taxonomic level.
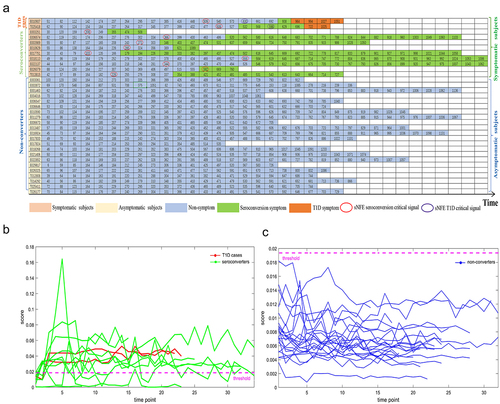
Figure 3. The curves of mNFE scores for all symptomatic subjects based on their respective DNBs at the overall taxonomic level.
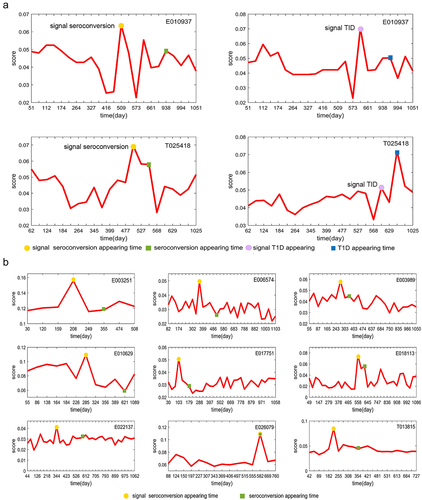
Table 1. Overview of the detection of early-warning signals based on the mNFE at different taxonomic levels.
Figure 4. Diversity analysis and accuracy analysis of mNFE.
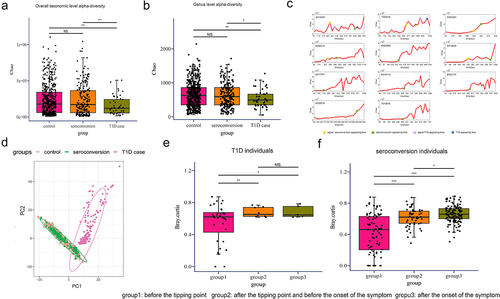
Figure 5. Functional analyses of high-frequency species.
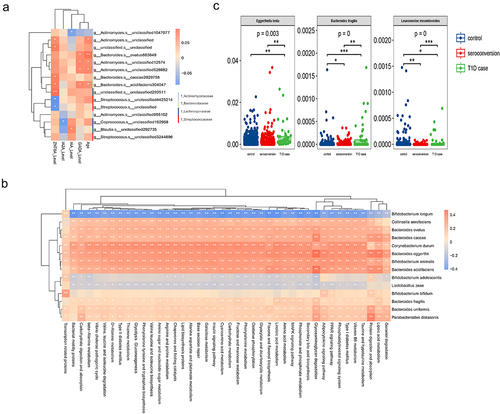
Figure 6. Analysis of the microbiome association network for each group.
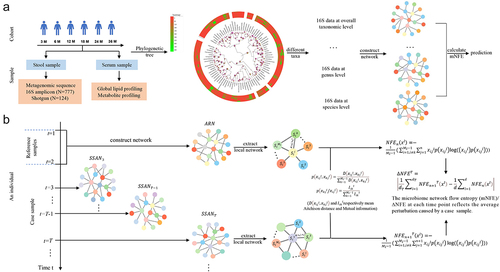
Figure 7. Schematic illustration of detecting the pre-disease state by using the association network based mNFE.