Figures & data
Figure 1. Assays investigation of deacetylation of CobB and its role in virulence regulation.
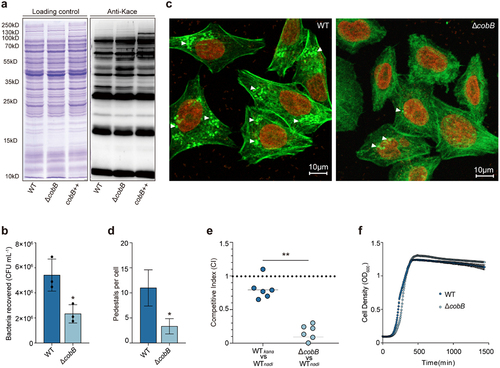
Figure 2. Identification of the substrates of CobB by using quantitative acetylome comparison of ΔcobB and WT.
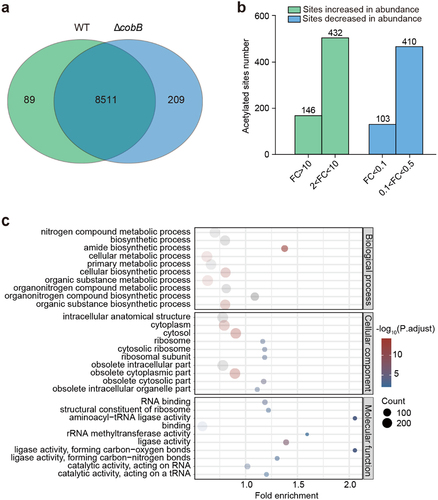
Figure 3. Assays of bacterial adhesion to HeLa cells and bacterial colonization of rabbits.
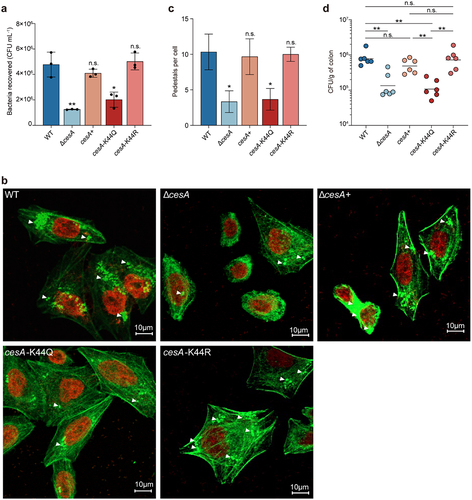
Figure 4. Assays of EspA stability and interaction with CesA.
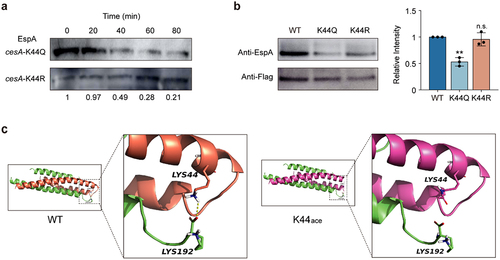
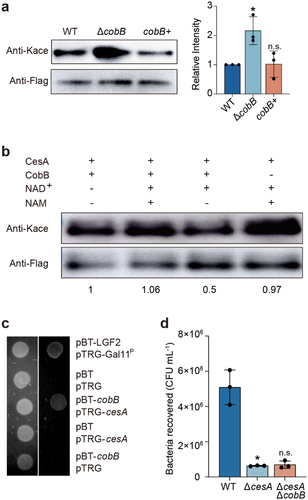
Figure 5. Assays of CesA interactions with CobB.
0311_Revised_Supplementary Information.docx
Download MS Word (25.6 KB)0311_Revised_Supplementary Datasets.xlsx
Download MS Excel (1.7 MB)Data availability statement
The relevant data are provided in the manuscript (Supplementary Information). MS proteomics data were deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD045950.
