Figures & data
Figure 1. Histopathological analysis of pulmonary and GI tissues in rhesus monkeys challenged with SARS-CoV-2 or variants.
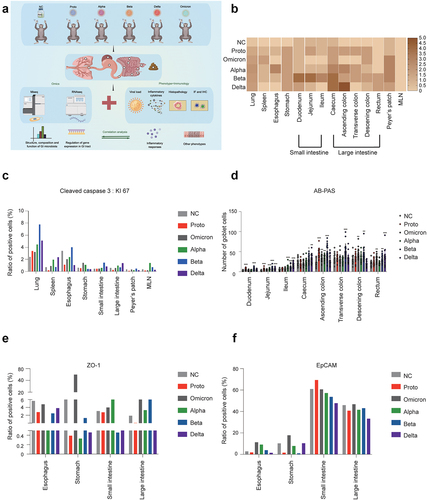
Figure 2. Inflammatory responses in GI tracts post SARS-CoV-2 proto strain or variants.
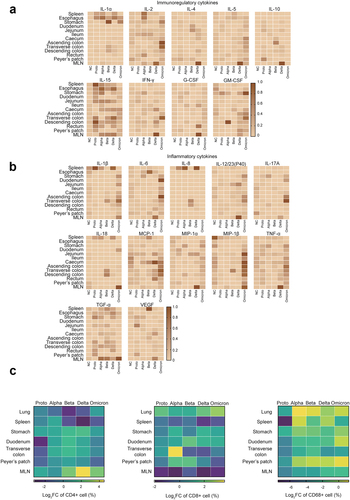
Figure 3. Microbial beta-diversity, composition and dominant microorganisms in GI contents of SARS-CoV-2 or variants infected animals.
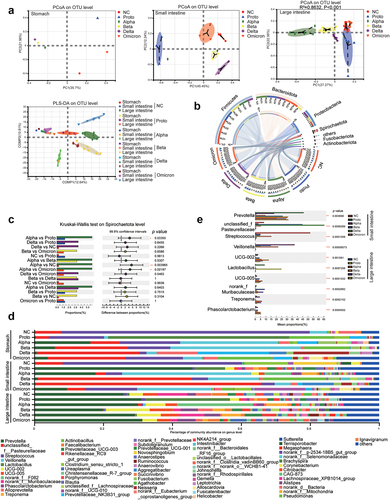
Figure 4. Prediction of microbial function and correlation analysis of tissue cytokines with microbiota in fragments of GI tract.
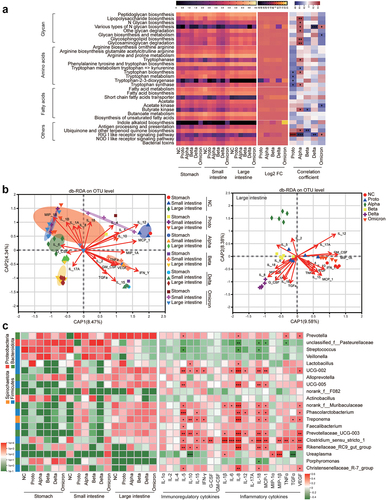
Figure 5. Transcriptomic profile of fragments in GI tract post SARS-CoV-2 or variants infection.
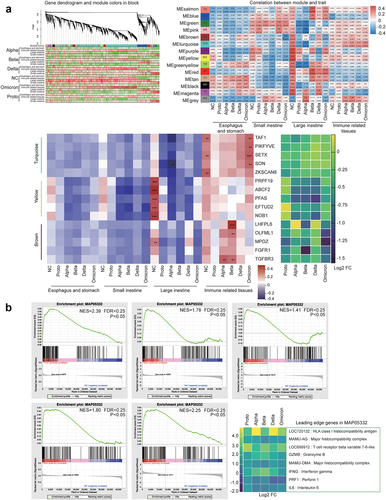
Table 1. Summary of pathogenic characteristics in two clusters of SARS-CoV-2.
