Figures & data
Figure 1. Schematic representation of four different groups of strategies for the treatment of C. difficile infections. Antibiotics, fecal microbiota transplantation (FMT), probiotics, and defined mixtures are discussed briefly in this review. The image was created with BioRender.
Figure 2. Selected TcdA and TcdB toxin-neutralizing antibodies and antibody alternatives. a. and b. Schematic representation and structural models of TcdA (merged from PDB IDs: 7POG and 2QJ6) and TcdB toxins (PDB ID: 6OQ5) with labeled binding sites of monoclonal antibodies actoxumab, bezlotoxumab, and PA41, the DLD-4 DARPin dimer, and small molecule compound ebselen. Toxin structures were visualized using ViewerLite 4.2 (Accelrys). GTD – glucosyltransferase domain, APD – autoprotease domain, DD – delivery domain, CROPS – combined repetitive oligopeptide sequences (receptor-binding domain). c. Schematic structures of various tandem VHH constructs (nanobodies). Individual nanobodies bind to distinct epitopes on the TcdA (light gray) or TcdB (dark gray) domains. Trx – thioredoxin, ABP – albumin-binding peptide.
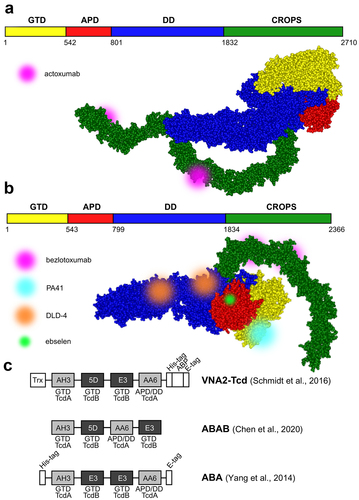
Table 1. Antibodies and antibody alternatives for neutralization of C. difficile toxins.
Table 2. Antibacterial peptides against C. difficile toxins.
Table 3. Agents for the neutralization of antibiotics in the colon lumen.
Table 4. Phage-based therapies with the ability to cause lysis of C. difficile..
Figure 3. Phage-based therapies against C. difficile infections. a. Conventional phage therapy: phages specifically dock to bacterial surface receptors (not shown) via tail fibers and inject their genomic DNA into host bacteria. Progeny phages are released upon bacterial wall disruption by phage-encoded endolysins which gain access to peptidoglycan through holin pores. b. Phages can be exploited as vehicles to deliver guide RNA (gRNA) which combines with the endogenous Cas nuclease of C. difficile to catalyze cleavage of the bacterial chromosome. c. Tailocins (phage tail-like particles) have no capsid head and therefore no phage genetic material. Nevertheless, they still selectively attach to bacterial surface receptors and puncture the cell, thereby disrupting the membrane potential. d. Recombinant phage endolysins degrade the peptidoglycan wall from the outside. The image was created with BioRender.
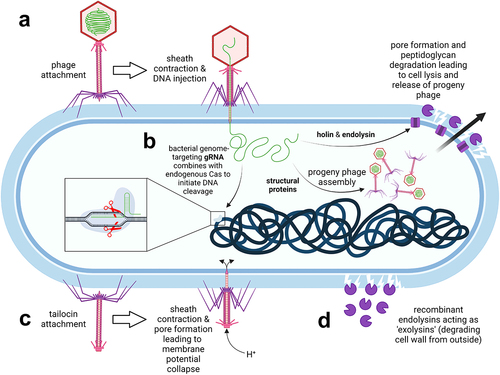
Table 5. Cytokines and anti-cytokine antibodies that facilitate clearance of C. difficile by modulating the immune system.
Table 6. Small-molecule agents against C. difficile that prevent the activity of C. difficile toxins, activate the immune system, affect bile acid synthesis, or exert their effects via other mechanisms.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
