Figures & data
Figure 1. Gut microbiota isolate whole-genome sequencing (WGS) similarity and Salmonella co-culture experimental workflow.
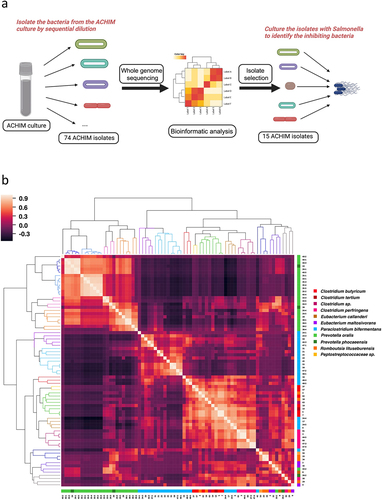
Figure 2. Gut microbiota isolates selectively reduce Salmonella growth.
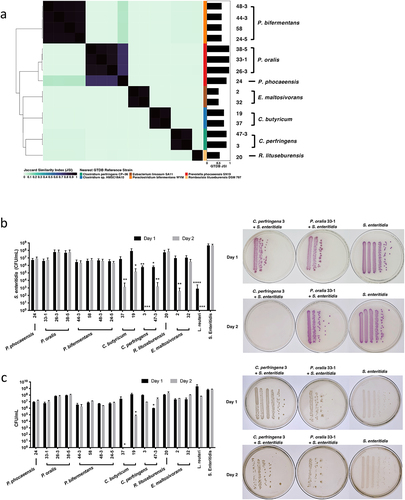
Figure 3. Evaluation of the functional genomic potentials among gut microbiota isolates.
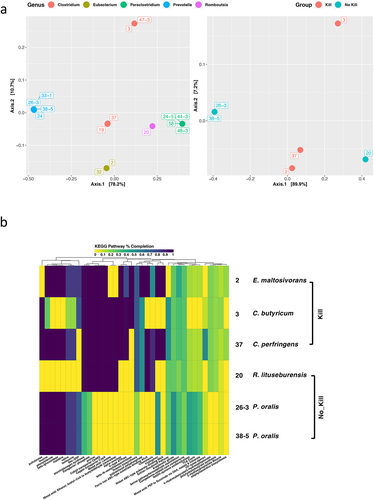
Figure 4. Metabolites unique in co-culture systems inhibiting Salmonella growth.
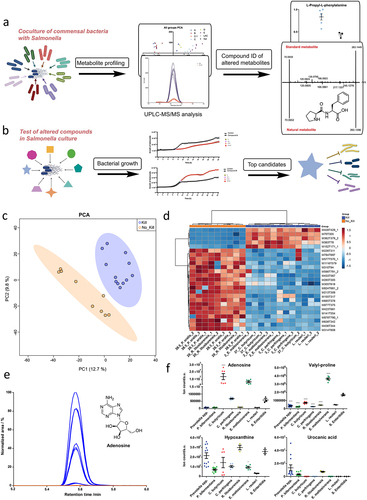
Figure 5. Adenosine and adenine are potent inhibitors of multiple-antibiotic-resistant pathogens.
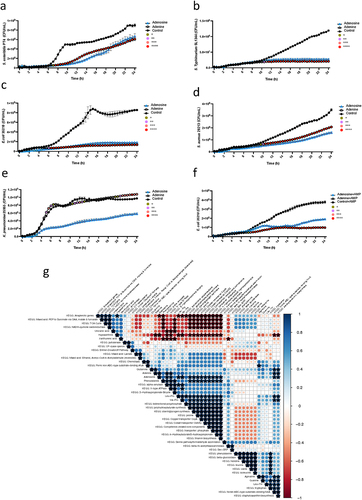
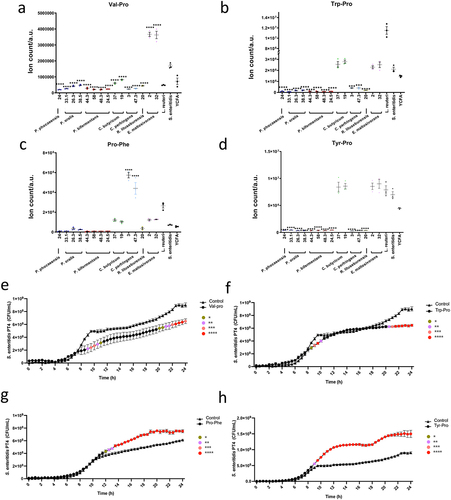
Figure 6. Dipeptides demonstrated the opposite function in Salmonella growth.