Figures & data
Figure 1. RstAB promotes LF82 virulence in chronic colitis mice.
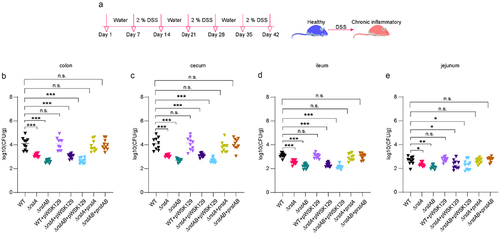
Figure 2. RstAB promotes the replication of LF82 in macrophages.
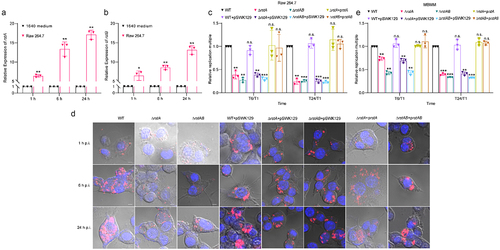
Figure 3. RstAB promotes LF82’s replication in macrophages by regulating the expression of csgD and asr.
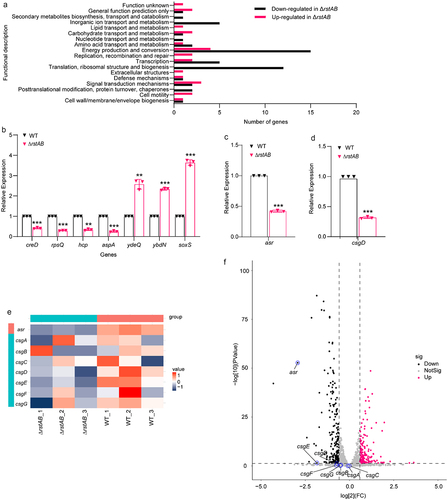
Figure 4. RstA regulates biofilm formation of LF82 by directly activates the expression of csgD.
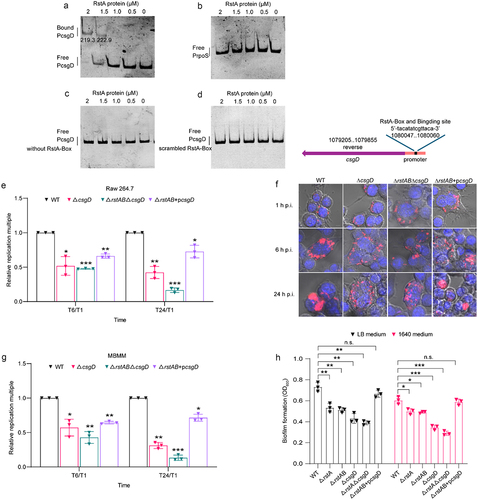
Figure 5. RstAB regulates the acid stress response of LF82 by directly activates the expression of asr.
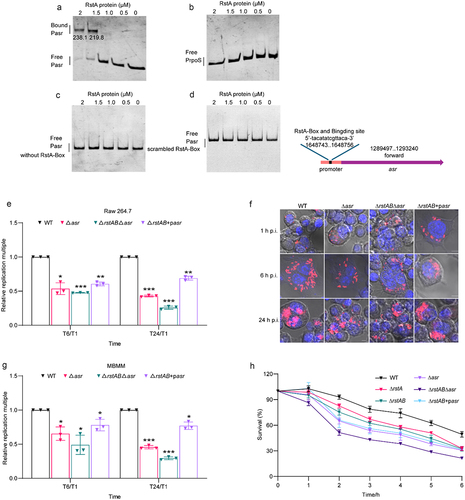
Figure 6. RstAB responds to acidic signal in macrophages and promotes intracellular replication of LF82.
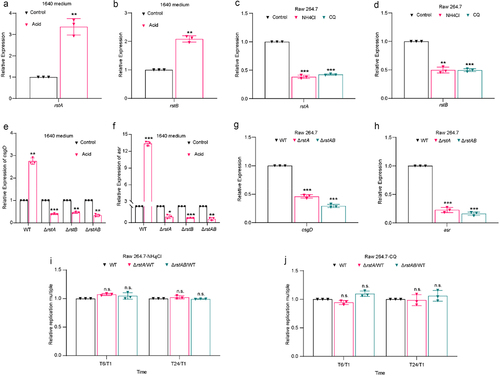
Supplemental Material
Download Zip (391.1 KB)Data availability statement
RNA sequencing data generated in this study are available from the NCBI SRA database. Accession to cite SRA data: PRJNA984413 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA984413.
