Figures & data
Figure 1. Tn-seq analysis for identification of functional genes under anaerobic conditions in E. faecium.
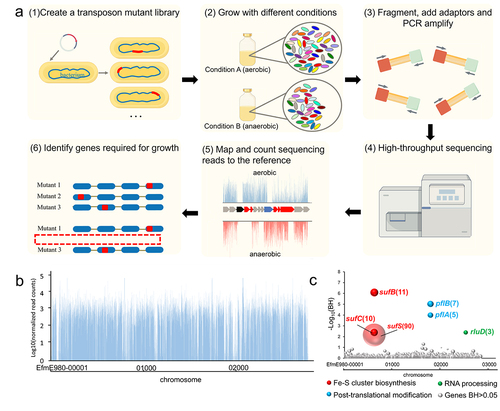
Figure 2. Essential gene of GO (a) and KEGG (b) enrichment analysis.
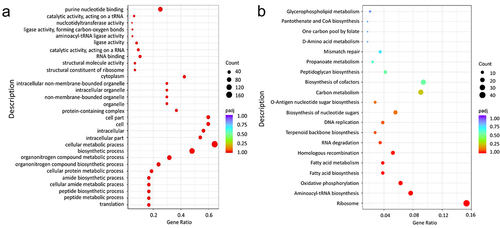
Figure 3. Phylogenetic and in silico analysis of Suf pathway.
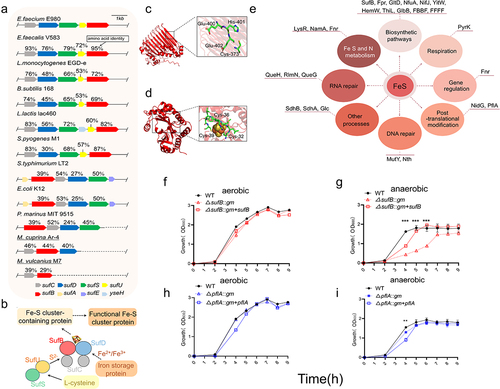
Figure 4. Transcriptome comparison between WT and sufB mutant strains under anaerobic conditions.
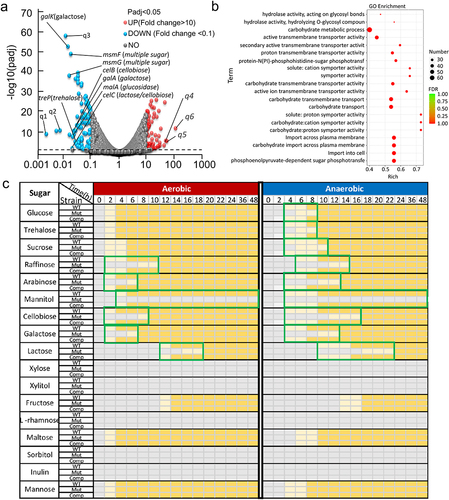
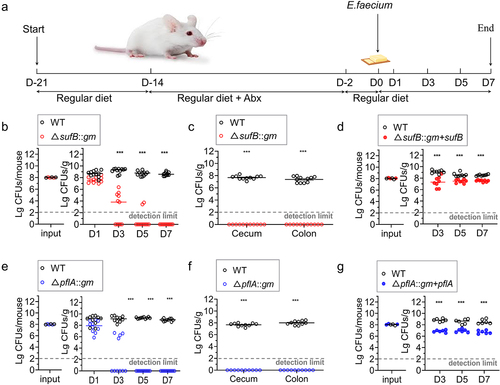
Figure 5. Colonization of intestine by E. faecium in a mouse model.
Supplemental Material
Download Zip (11.3 MB)Data availability statement
The datasets generated during this study are available in the National Center for Biotechnology Information (NCBI) BioProject Repository https://www.ncbi.nlm.nih.gov/bioproject under BioProject PRJNA1061835.
