Figures & data
Figure 1. The myosin IQ motif in the heavy chain. (A) Schematic representation of a generic myosin heavy chain. The N-terminus of the heavy chain represents the catalytic motor domain (gray) that harbors the nucleotide binding site and the F-actin binding region. The central neck domain harbors the IQ motifs (orange). The C-terminal tail domain (dark gray) can contain a vast collection of domains and motifs that determine the oligomeric state of the heavy chain and interaction signatures with binding partners. The length of the myosin motor domain is very conserved, whereas the length of the myosin neck and tail domain varies considerably. Some myosins have an N-terminal extension of variable length prior to the motor domain which is indicated by the dashed line. (B) The top line represents the consensus IQ motif found in myosin heavy chains. Position 1 of the 11 amino acid long consensus IQ motif is critically occupied by a hydrophobic residue such as isoleucine (I), leucine (L) or valine (V) though methionine (M), phenylalanine (F) and lysine (K) and threonine (T) are found in some cases. Position 7 is ambiguous for several amino acids. Position 11 is preferentially occupied by a positively charged amino acid (arginine (R), histidine (H) or lysine (K)). Invariant residues are boxed in gray, ambiguous residues in peach. Non-boxed residues are highly variable. The WXW motif found in the hook region of conventional myosins-2 is colored in rose. Of note, the WXW motif is not part of the consensus IQ motif but crucial for the interaction of RLCs with the IQ2 in myosins-2 and therefore depicted for completeness. All other lines shown represent selected IQ motif found in conventional and unconventional myosin heavy chains discussed in this review. The abbreviations used are as follows: M1: IQ1 and IQ3 from human myosin-1C, M2: IQ1 and IQ2 from chicken striated muscle myosin-2; M5: IQ2 and IQ4 from budding yeast Myo2p; M6: IQ1 from pig myosin-6; M7: IQ3 from human myosin-7A; M10: IQ3 from human myosin-10; M14: IQ2 from P.yoelli. (C) Crystal structure of a prototypic myosin-2–2IQ fragment. Structural overview of the striated myosin-2–2IQ fragment from scallop (PDB ID: 1DFL). The myosin heavy chain is shown in gray cartoon representation, the nucleotide in black spheres. The associated ELC and RLC are represented in orange and cherry color, respectively. ELC and RLC bind distal from the globular motor domain to the IQ motifs of the neck domain. The myosin neck region and the associated light chains form the lever arm. The hook is indicated by a gray arrow.
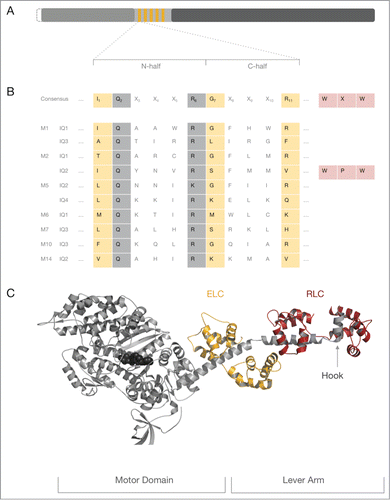
Table 1. List of the commonly used historical and recent abbreviation for ELCs, RLCs and CaM in different organisms as found in the NCBI database
Table 2. List of selected myosin heavy chains and identified light chains discussed in this review
Figure 2. Structure of CaM and its EF-hand motifs in various conformations. (A) Structures of CaM (blue) bound to a target sequence (gray) in the extended (PDB ID: 3CLN) and compact conformation (PDB ID: 2IX7). (B) EF-hand conformations found in CaM. Top row: EF-hand in an open (PDB ID: 1CLL) or closed (PDB ID: 1CFC) conformation. Bottom row: The semi-open (PDB ID 2IX7) and the uncoupled conformation of the EF-hand motif (PDB ID 4R8G). is adapted from Lu et al.Citation34
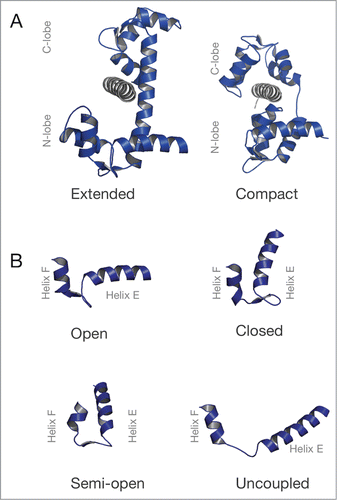
Figure 3. Structural consequences of RLC phosphorylation on the conformation of smooth and nonmuscle myosins-2. RLC phosphorylation at S19 promotes a conformational change from a compact (left) to an extended conformation of the myosin holoenzyme (left), which readily assembles into higher order bipolar filaments (not shown). The structural transition from the compact to the expended conformation is accompanied with an increase in the mechanoenzymatic activity. RLC dephosphorylation by myosin phosphatase reverses the conformational change. Figure adapted from Heissler and Manstein.Citation87
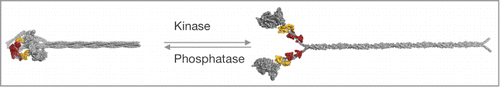
Table 3. Compilation of selected myosin and non-myosin binding partners of ELC and RLC. It is of note that not all kinases and potentially phosphatases that interact with the RLC are listed due to space limitations
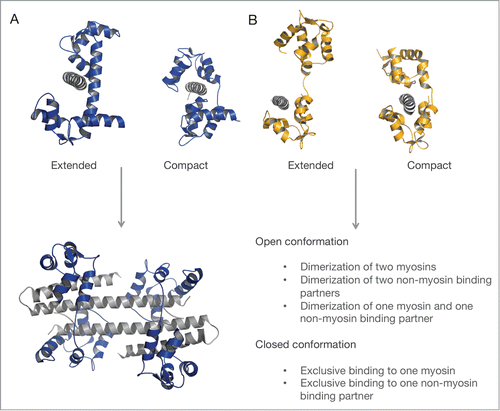
Figure 4. Consequences of different binding modes of myosin light chains. (A) Schematic representation of the open and closed conformation of CaM (blue) bound to an exclusive target IQ-motif (gray). CaM-dependent dimerization of 2 domains of the Ca2+-activated K+ channel (gray), each bound to one lobe of CaM (PDB ID: 1G4Y). (B) Extended and compact light chain conformation exemplarily shown for the ELC (orange) on the IQ motifs (gray) of the yeast myosin-5 (PDB IDs: 1M45, 1M46). Potential functional consequences of the extended and compact light chain conformation are valid for ELC, RLC and CaM.