Figures & data
Figure 1. Functional consequences of silencing LMNB1. (A) Immunoblot showing decreased levels of LMNB1 in three single cell subclones (shown in duplicates). Note that in two out of three clones no major disruption of the NL was observed when assessed the expression of lamin B2 and lamin A/C. GAPDH was used as loading control. (B) Cell viability analysis of LMNB1-depleted cells after silencing LMNB2 and LMNA by siRNA assessed at 96 hours after transfection. AllStar Death was used as positive control.
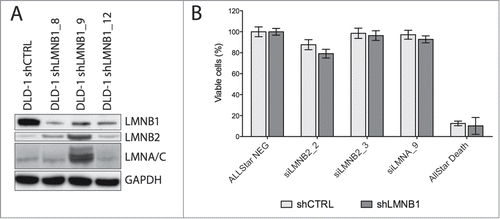
Figure 2. Role of LMNB1 in the regulation of the chromosome architecture and DNA transcriptional activity. (A) 3D-FISH showing chromosome 18 labeled in Spectrum Orange and chromosome 19 in FITC in control and LMNB1-depleted DLD-1 cells. Bar, 5 μm. (B-C) Plots indicating the increase of both volume and surface of chromosome territories for HSA18 and HSA19 in the LMNB1-depleted cells. Measurements of the digitalized chromosome territories were taken to determine changes in volume and surface. (D) Scattered distribution of the histone mark H3K27me3 in LMNB1-depleted cells compare to the peripheral localization in control cells. Bar, 2 μm. (E) Quantification of the signal distribution of the epigenetic histone mark H3K27me3 determined by using Image J software (n = 600). (F) Functional GO terms significantly deregulated associated with transcriptionally altered spliced genes as a consequence of LMNB1 silencing.
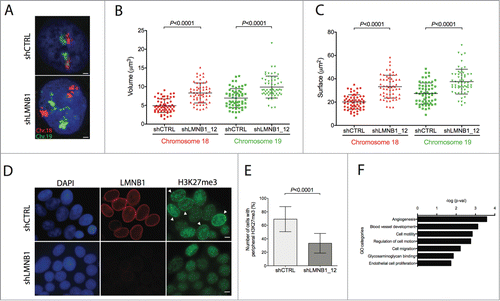
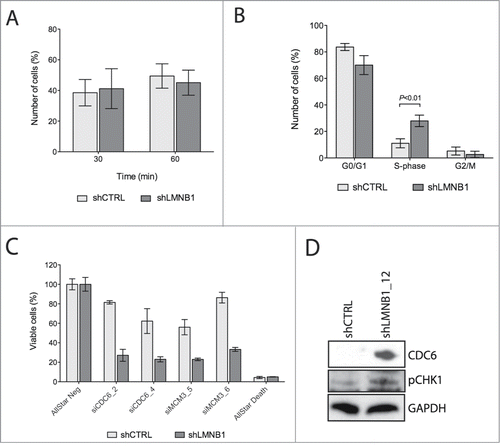
Figure 3. LMNB1 cooperates with the formation of DNA pre-replication complexes. (A) Cells were incubated with EdU for 30 or 60 minutes, respectively. After EdU incorporation cells were fixed and stained. EdU positive cells were counted (n > 400 ). (B) FACS analysis showing differences in cell cycle stages in shCTRL and shLMNB1 cells. (C) Cellular viability after LMNB1-depleted cells were transfected with siRNA against CDC6 and MCM3. (D) Immunoblot for CDC6 and the phosphorylated form of CHK1 showing an increase amount of these proteins when LMNB1 is silenced.