Figures & data
Figure 1. Normal and anomalous diffusion. Typical areas covered by 2 telomeres that undergo normal diffusion (A) and anomalous diffusion (B) over 50 time-points acquired during 925 seconds. (C, D) Trajectories of telomeres (green spots) and Cajal bodies (red spots) acquired from time-lapse fluorescence live-cell imaging over 925 seconds in Lmna−/− (C) and Lmna+/+ (D) cells. Note that the volume of movement in the Lmna−/− cells is much larger compared to that in Lmna+/+cells. Scale bar, 3µm.
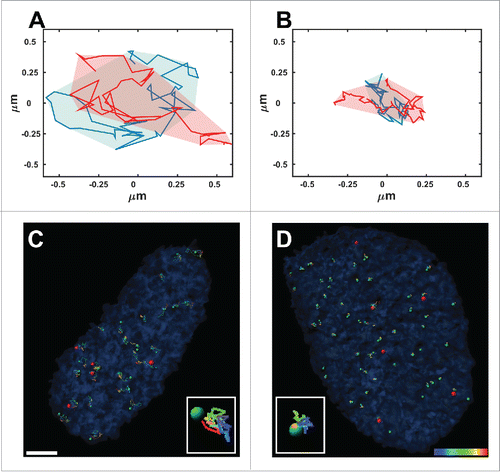
Figure 2. Diffusion patterns of telomeres in different cell types. (A) MSD/Δt vs Δt plot in log-log scale for different cell lines. (B) The distribution of α coefficient obtained from single telomeres trajectories (n=325, 166, 551, 958 for the 3T3, HeLa, MF, U2OS cell-lines respectively). (C) MSD/Δt vs Δt plot for Lmna+/+and Lmna−/− cells. (D) Box plot for the α coefficients distribution (n=223, 525 for the Lmna+/+ and Lmna−/− cells respectively). Note the significant difference showing anomalous diffusion in Lmna+/+ and normal diffusion in Lmna−/− cells.
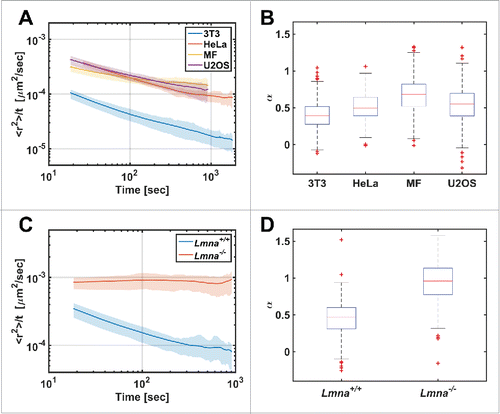
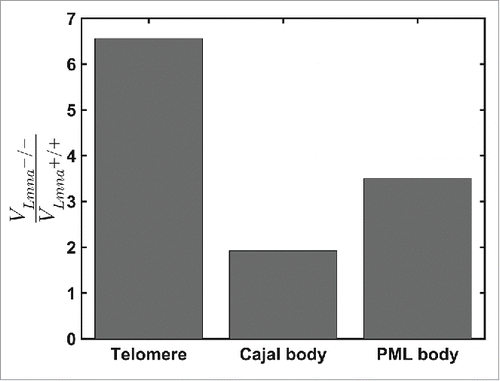
Figure 3. Comparison of the ratio of volumes of motions for telomeres, Cajal and PML bodies during 925 seconds in Lmna+/+ and Lmna−/− cells. For Lmna+/+ cells n=691, 74, 78 for telomeres, Cajal bodies and PML bodies, respectively. For Lmna−/− cells n=519, 109, 76 for telomeres, Cajal bodies and PML bodies, respectively. All entities explore smaller volumes in the Lmna+/+ compared to the Lmna−/− cells. The effect of lamin A depletion is more significant on chromatin dynamics (telomeres) compared to nuclear bodies.