Figures & data
Figure 1. Ubiquitin-proteasome-dependent protein degradation pathways in the yeast nucleus. San1 is a nuclear E3 protein ubiquitin ligase that ubiquitinylates misfolded nuclear (NP) and cytoplasmic (CP) proteins. Delivery of CPs to nuclear San1 is assisted by Hsp70 chaperone Ssa1. Unlike the ER-membrane localized E3 ligase Hrd1, E3 ligase Doa10 localizes to both ER and the INM and targets INM protein Asi2 and transcriptional repressor Matα2 for proteasomal degradation. Asi1-Asi3 is an E3 ligase complex enriched in the INM that, together with Asi2, ubiquitinylates latent forms of transcription factors Stp1 and Stp2 via their RI degron. Asi1-Asi3 also ubiquitinylates misfolded or mislocalized integral membrane proteins (IMP) in the INM. The nucleus is also the compartment for the proteotoxic stress-induced deposit of misfolded cytoplasmic proteins and protein aggregates in the intranuclear quality control compartment (INQ).
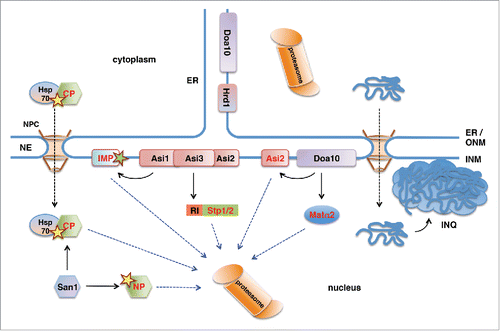
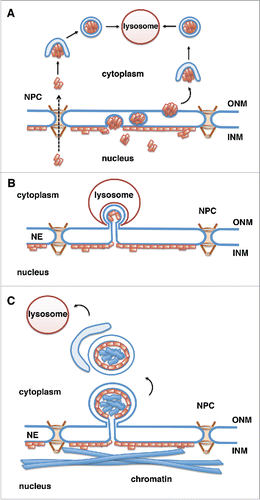
Figure 2. Potential mechanisms for lysosomal degradation of progerin by autophagy. Progerin (depicted by orange rectangles) is a lamin A mutant that associates with the INM. Progerin may become accessible for degradation by the cytoplasmic lysosomes in the following ways. (A) Progerin aggregates translocate to the cytoplasm through nuclear pores (NPCs) or vesicle-mediated transport through the double membrane of the NE (nuclear egress). Progerin aggregates are engulfed by the autophagosomal membrane in the cytoplasm and fuse with the lysosome.Citation67 (B) Similar to piecemeal microautophagy of the nucleus in yeast, blebs of the nuclear envelope are engulfed by invaginations of the lysosomal membrane and then pinch off into the lysosomal lumen. (C) Vesicles or micronuclei bud off from the nuclear envelope, are engulfed by the growing isolation membrane producing autophagosomes, which fuse with lysosomes and deliver nuclear components to the lysosomal lumen.