Figures & data
Figure 1. The importance of fork protection in maintaining genome stability: i. Cellular replication forks can stall for a variety of reasons. In certain circumstances, stalled forks may reverse to aid repair or restart. ii. A number of fork protection factors including RAD51 (described in the text) act to protect nascent DNA from over-processing by cellular nucleases (denoted by ‘pacman’ symbols). iii. This allows subsequent fork restart and/or repair by homologous recombination, and prevents genome instability. iv. In the absence of these protective factors, excessive nucleolytic processing of stalled/reversed forks leads to common fragile site and chromosomal instability, or to inappropriate repair giving rise to chromosomal fusions and radial chromosomes, ultimately leading to genomic instability (v).
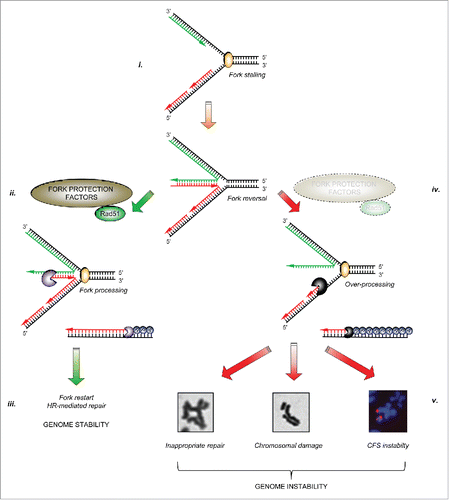
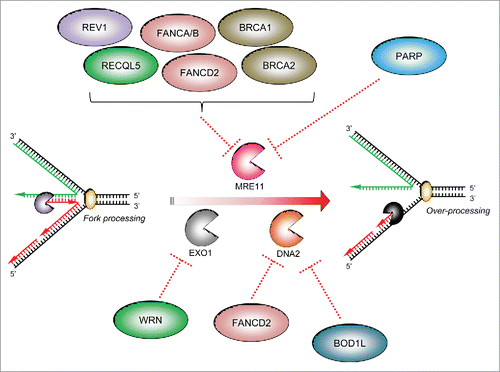
Figure 2. The specificity of factors counteracting ‘fork’ nucleases: Three main cellular nucleases have thus far been implicated in the over-processing of stalled forks: MRE11, DNA2 and EXO1. There have been no reports that the nuclease CtIP is involved in over-processing of stalled replication forks. Several protective factors act on specific cellular nucleases to supress their aberrant activity on stalled replication forks (dotted red lines): several FA/HR proteins, the TLS polymerase REV1 and PARP1 have all been reported to inhibit MRE11-dependent fork resection, whilst the WRN helicase/nuclease prevents EXO1-dependent fork degradation. Recently, we demonstrated that BOD1L is required to suppress DNA2-dependent strand degradation of stalled replication forks, alongside speculative reports of a similar role for FANCD2.