Figures & data
Figure 1. Depletion of LINC complex components caused morphological changes in cells. (A) MCF10A cells were transfected with the indicated siRNAs and stained with anti-SUN1 (left), -SUN2 (middle), and -lamin A/C (right) pAbs, respectively, to show that each knockdown was efficient. Cell nuclei were counterstained with DAPI. Scale bar = 20 µm. (B) Pap-stained images of the cells transfected with the siRNAs. The rectangles in the top panels indicate regions enlarged at the bottom. Scale bar = 20 µm. (C) Classification accuracy (CA) of knockdown against control (siControl_1) cells was measured using the machine-learning algorithm wndchrm. Twenty images for each knockdown class (Fig. S2) were employed for the CA measurements. Knockdown of SUN1, SUN2, and lamin A/C resulted in morphological changes significant enough to yield high classification accuracy (CA= ∼1.0 for each). The dotted line indicates CA = 0.5, the expected value of random classification with no detectable morphological differences. Thus, the value for the difference between siControls_2 and _1 (CA = 0.462 ± 0.083) is approximately as expected. (D) Morphological distances of the knockdown classes from the control were measured using wndchrm. Twenty images for each knockdown class (Fig. S2) were analyzed. Larger values indicate morphologies different from those of control cells (siControl_1). Knockdown of SUN1 resulted in significant changes. For (C) and (D), the values are the mean ± standard deviation (SD) of 20 independent cross validation tests. p values were calculated using the Student t test (***p < 0.005).
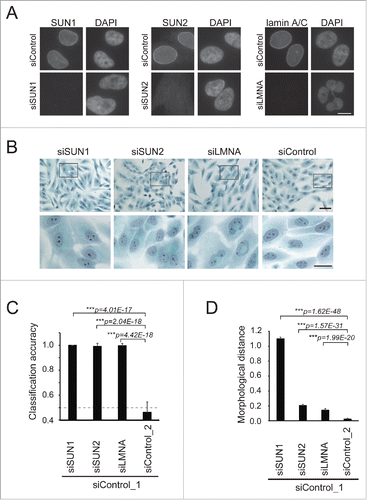
Figure 2. Depletion of SUN1 alters nucleolar structure. (A) Representative images for nuclear regions of the knockdown cells extracted from the Pap-stained images. Scale bar = 25 µm. (B) Morphological distances of the knockdown classes from the control (siControl_1) were measured using wndchrm. Sixty images (Fig. S3) were applied. Knockdown of SUN1 resulted in significant changes in the nucleus. (C) Fisher discriminant scores assigned for each test in . Fisher scores were computed for 4,008 features that were grouped as follows: ChFT, Chebyshev-Fourier transform; Ch, Chebyshev Statistics; Cf4M, Combined First Four Moments; Fra, Fractal Statistics; Har, Haralick Texture; MSH, Multiscale Histogram; Rad, Radon; Zer, Zernike; etc, others. Fisher score values for each individual feature are listed in Supplementary Table S2. In the bottom four panels, relative scores were calculated from the top panels. The maximum Fisher score in each test was defined as 1. (D) The region corresponding to the nucleolus (left, red box) was extracted from the Pap-stained images. If the cells contained more than two nucleoli, all were extracted. Representative images for each class are shown. Scale bar = 2 µm. (E) Morphological dissimilarities of the knockdown classes from control (siControl_1) were measured using wndchrm. Seventy-five images (Fig. S4) were applied. Knockdown of SUN1 resulted in significant changes in the nucleolus. For (B) and (E), the values represent the mean ± SD of 20 independent cross-validation tests. P values were calculated using the Student t test (***p < 0.005, n.s. not significant).
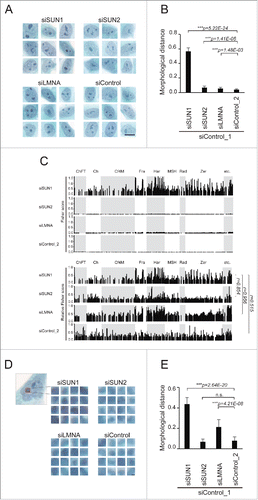
Figure 3. The larger and reduced number of nucleoli were formed in the SUN1-depleted nucleus. (A-C) Immunofluorescence analysis of SUN1-depleted MCF10A cells. Subnucleolar regions of FC, DFC, and GC were visualized using anti-UBF (A), anti-FBL (B), and anti-NPM antibodies (C), respectively (green), and chromatin DNA was counterstained with DAPI (red). Scale bar = 10 µm. (D-G) Quantitation of FBL-labeled nucleolar regions. Area of each nucleolus (D), number of nucleoli per nucleus (E), total nucleolar area per nucleus (F), average signal intensity of the nucleolus (G). The values represent the means ± SD (n >25 cells). P values were calculated using the Student t test (**p < 0.01, n.s. not significant). For (D), (F) and (G), the relative values are shown in arbitrary units (a.u.). The value for the siControl was defined as 1. (H) Gallery of nuclear images used for analysis in . DAPI-stained nuclear regions of SUN1-depleted MCF10A cells. Bar, 10 µm. (I) Morphological distance of knockdown classes from the control (siControl_1) were measured using wndchrm. The values represent the mean ± SD of 20 independent cross-validation tests. P values were calculated using the Student t test (***p < 0.005). (J) The dendrogram reflects the morphological similarity between siControl- and siSUN1-transfected cells. The dendrogram was constructed from 20 cross-validation tests using wndchrm (n = 13 images).
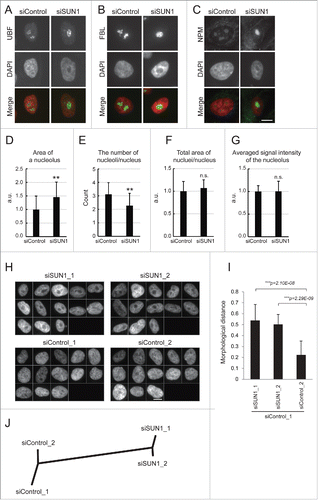
Figure 4. Depleting SUN1 suppresses rRNA synthesis but not rRNA processing. (A) The levels of pre-rRNAs in the indicated MCF10A cells were quantitated using real-time PCR with primers specific for the 5′ external transcribed spacer region of pre-rRNA. Values represent the mean ± SD for triplicates. (B) Cellular RNAs were pulse-labeled with 3H-UTP for 2 h and chased with nonradioactive UTP for the indicated period. Total RNAs were isolated and separated on the agarose gel. Positions of the nascent (47S/45S), intermediate (32S) and 28S and 18S rRNAs are indicated next to the gel.
Figure 5. Nucleolar size correlates negatively with SUN1 expression levels in human breast cancer tissues. (A) Breast tumor specimens from patients were stained using anti-SUN1 pAbs (brown), and cells were counterstained with hematoxylin (blue). A representative example both noncancerous and cancerous regions is shown. The left part of this panel shows the cancer-associated noncancerous region maintaining persistent ducts, and the majority of the cells were SUN1 positive. The right part of the panel shows cancerous regions with reduced SUN1 staining. Scale bar = 50 µm. Representative images of SUN1-positive cells with small nucleoli (a) and SUN1-negative cells with enlarged nucleoli (b) are enlarged at the bottom. The areas of the nucleoli in SUN1-positive or in SUN1-negative cells were 118 ± 39 and 37 ± 19, respectively (n >10 cells). (B) Cancer-associated non-cancerous mammary ducts composed of heterogenous population of cells are shown. Small inflammatory cells surround the SUN1-positive mammary duct. Scale bar = 50 µm. A cell with strong SUN1-staining (a) localized to the periphery of the duct and has small nucleolus, while one with faint SUN1-staining (b) is inside of the duct and harbors a large nucleolus.
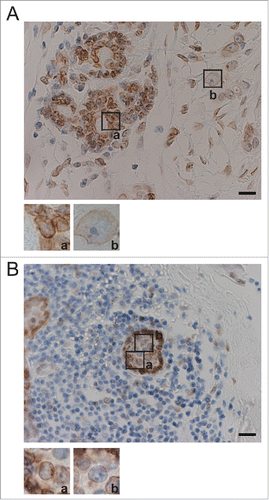
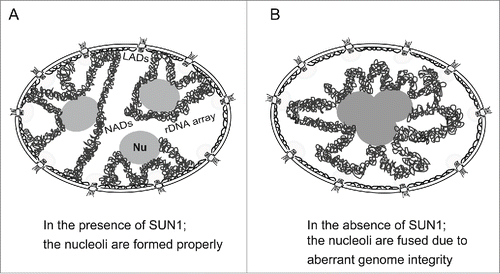
Figure 6. Proposed model for nucleolar fusion upon depleting SUN1. (A) Nucleoli are properly formed in the presence of chromatin associated with the nuclear membrane through SUN1. (B) The nucleoli are fused in the absence of chromatin anchoring at the NE through SUN1. Nu, the nucleolus; LADs, lamina-associated domains; NADs, nucleolus-associated chromatin domains.