Figures & data
Figure 1. Genetic instability generated by DSB repair. (A) Two types of genetic instability generated by DSB repair: error-prone end-joining resulting in mutations at the junctions (left panel) and rearrangements resulting from the joining of distant DSB (right panels). Mutagenic joining and rearrangements can be associated (right panel). (B) Intra-chromosomal substrate monitoring end-joining. Reporter genes (GFP or CD4 membrane antigen) are not expressed because of an intervening sequence (in blue) between the reporter gene and promoter. Two cleavage sites for the meganuclease I-SceI (red arrows) are inserted in the intervening sequence. Expression of I-SceI excises the intervening sequence and end-joining places the reporter gene under the control of the promoter. The expression of the reporter genes enables monitoring of end-joining events, and sequencing of the repair junctions enables monitoring of the repair accuracy. (C) Two-ended DSB (left and middle panels) vs. one-ended DSB induced by replication fork arrest (right panel). The tethering (gray spheres) maintains genome organization. With two distant DSBs (middle panels), bypass of the tethering leads to genetic rearrangements. Intermolecular end-joining (upper middle panel) result in translocation. Intramolecular end-joining (lower middle panel) result in either deletion or inversion of the intervening sequence. The joining of one-ended DSBs (right panel) inevitably leads to genetic rearrangements.
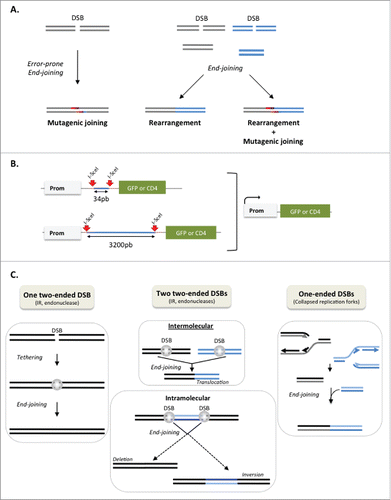
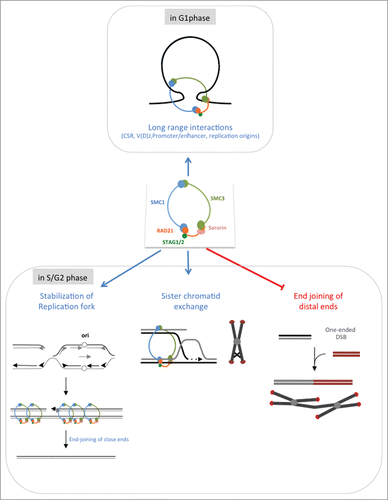
Figure 2. Multiple roles for the cohesion complex. In the G1 phase (upper panels). In the S/G2 phase (lower panel): impact of sister chromatid cohesion in genome stability maintenance following replication stress. Arrested replication fork are stabilized by the cohesin complex (left panel). A converging migrating replication fork reaches the blocked fork, leading to the formation of 2 close ends. The cohesin complex does not prevent the joining of such DSEs. Moreover, the end-joining of close ends is more conservative (Guirouilh-Barbat et al., submitted). Alternatively, sister chromatid exchange (SCE) enables the arrested replication fork to resume (middle panel); because the sister chromatids are identical, this process favors genetic stability. In the absence of the cohesin complex (right panel), one one-ended DSB can interact with another distant DSE. The joining of such DSEs generates chromosome fusions and genome rearrangements. The cohesin complex not only favors the 2 former situations, which both maintain genome stability but also restrains the joining of distant ends, protecting against the associated genetic instability.