Figures & data
Figure 1. Replication fork proteins of Saccharomyces cerevisiae. The Table to the left lists the names of subunits, complexes, their molecular weight and biochemical function. SDS polyacrylamide gels of recombinant pure proteins are shown to the right of the Table.
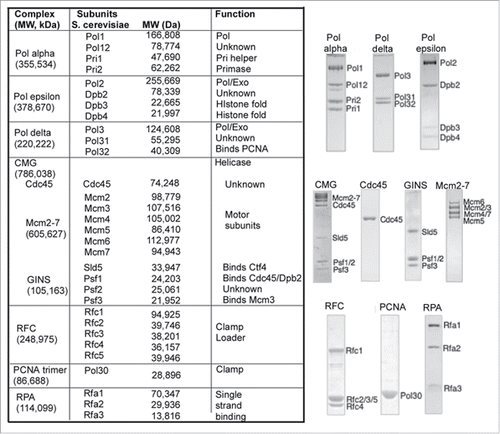
Figure 2. Example of leading-lagging strand replication in vitro. (A) The substrate used in the in vitro assays is a 3 kb forked DNA that has no dG residues on the leading strand and no dC residues on the lagging strand. Therefore 32P-dCTP specifically labels the leading strand product and 32P-dGTP labels lagging strand products. (B) Leading strand reaction time course using 32P-dCTP and either Pols α + epsilon, or Pols α + epsilon + delta. (C) While a low level of lagging strand products are observed with only Pols α + epsilon, significantly more lagging strand fragments are formed by addition of Pol delta. (D) Quantitation of DNA synthesis shows similar extents of leading and lagging strand replication. The panels of the figure are adapted with permission fromCitation50. Specifically, panels a, b are from Supplemental Figure 6a,b, panel c is from Figure 5b, and panel d is from Figure 7b of Citation50.
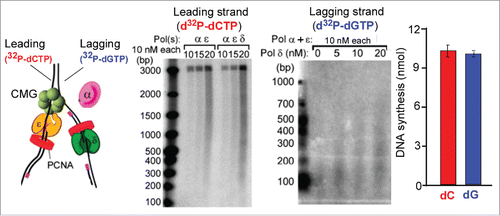
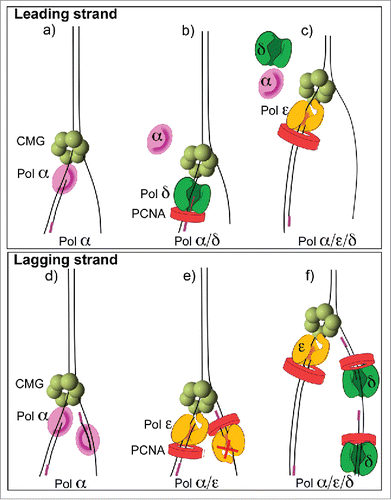
Figure 3. Asymmetric use of DNA polymerases at a replication fork. The diagrams refer to conclusions from in vitro replisome reconstitution reactions demonstrating that Pol epsilon is the dominant enzyme on the leading strand and Pol delta is the dominant enzyme on the lagging strand Citation38,43. (A) Pol α functions with CMG on the leading strand, (B) Pol delta switches with Pol α on the leading strand but lowers synthesis, (C) Pol epsilon takes over from both Pols α and delta to provide the most synthesis on the leading strand. (D) Pol α can function on the lagging strand in the absence of other polyemrases, (E) Pol epsilon takes over from Pol α on both strands, but is only active on the leading strand, (F) Pol delta extends Okazaki fragments in the presence of Pol epsilon and Pol α.