Figures & data
Figure 1. Remosome model resulting from a molecular simulation. This snapshot presents the DNA loop generated by a 42 bp insert at the dyad position.Citation17 The image shows the solvent-excluded surfaces of the histone core (gray) and of the DNA (cyan, partially transparent) as well as the helical axis of the DNA (black). Note the sharp kink within the DNA loop on the left hand side of the image. Molecular graphics were generated with the UCSF Chimera package.Citation27
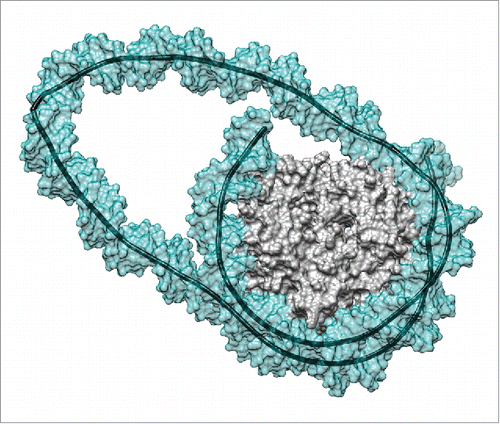
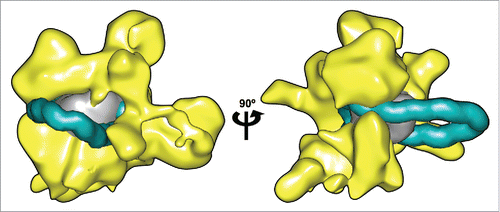
Figure 2. Remosome model placed within the RSC remodeller. Two views of a nucleosome containing a 42 bp insertions at the dyad (DNA shown in cyan, histone core in gray) manually docked inside the 3D reconstruction of the RSC complex calculated from cryo-EM data.Citation3 (shown in yellow). A representative snapshot of the remosome model was taken from a 0.5 μs MD simulationCitation17 converted to a 25 Å-resolution density map to match the RSC data. DNA in the looped nucleosome is shown up to 25 bp from the ends, to account for the possible rearrangements in these zones.Citation3 Molecular graphics were generated with the UCSF Chimera package.Citation27,28