Figures & data
Figure 1. Structure and expression of SUN1 splice variants. (A) A comparison of SUN1 splice variants, including a novel variant, SUN1_888. The SUN1 gene contains 22 exons, which are indicated by black boxes. Lines are introns. Exons 6−9 are alternatively spliced to generate numerous SUN1 variants. SUN1 variant, SUN1–001, composed of 822 aa (ensemble SUN1–001), UniprotKB-O94901 variant composed of 812 aa (KIAA0810, Nagase et al., 1998), SUN1 variant1 composed of 785 aa (NM_001130965), and testis specific SUN1 variant2 composed of 702 aa (NM_025154), are here referred to as SUN1_822, SUN1_812, SUN1_785, and SUN1_702, respectively. (B) Schematic diagram of SUN1_888, SUN1_785, and SUN1_916 proteins and amino acid sequence alignment of the variable region of SUN1 proteins. Human SUN1 proteins are single pass type II transmembrane proteins of the INM with an approximately 450 amino acid C-terminal region extending into the perinuclear space (PNS), which includes the SUN domain, and an approximately 200−400 amino acid N-terminal region extending into the nucleoplasm.Citation1,33 Identical residues in the amino- and carboxy-regions are shown as solid lines. Functional domains are indicated; the transmembrane domain (TM) and the lamin A/C binding site are retained in all of these variants. SUN domains are boxed.Citation1,5 (C) Expression of various SUN1 splice variants in human tissues was analyzed by reverse transcription PCR (RT-PCR). The amplicons were analyzed in 6% TBE polyacrylamide gels. Similar to mouse SUN1,Citation34 the shortest SUN1 variant, SUN1_702, could be detected exclusively in testis but not in any somatic tissues tested. * corresponds to a non-specific amplicon, as judged by direct sequencing and SUN1 knockdown (data not shown). Exon 7-, 8-, or 6-deleted SUN1 variants were not found in any database but we detected corresponding bands (indicated as ***) in various tissues, including the brain, heart and skeletal muscle. (D and E) HeLa cells were lysed with RIPA buffer and immunoprecipitated with indicated antibodies. Then input lysate (D) or precipitated proteins (E) were analyzed by western blotting using anti-SUN1 pAb.
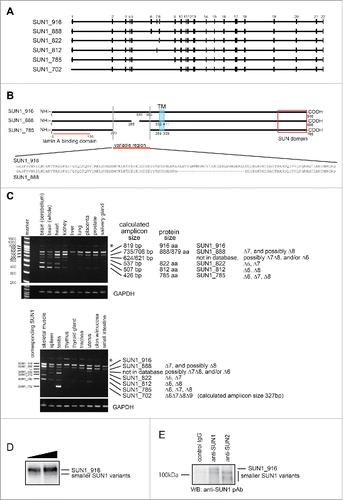
Figure 2. SUN1 and SUN2 are essential for cell migration. (A) MDA-MB-231 cells were transfected with siRNA pools that specifically target SUN1 or SUN2, termed siSUN1 or siSUN2, respectively, or with non-targeting siRNA as a negative control. Forty-eight hours later, total cell lysates were analyzed by western blotting using anti-SUN1 or anti-SUN2 pAbs. (B) Forty-eight hours after SUN1 and/or SUN2 knockdown, cell migration activities were analyzed. Each bar represents the mean number of cells per field ± SD. *; p < 0.05, **; p < 0.01.
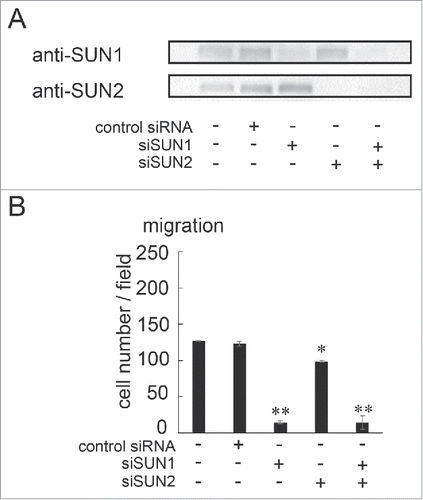
Figure 3. overexpression of SUN1_785 and SUN1_888 but not SUN1_916 promotes cell migration activities. (A) MDA-MB-231 cells were transfected with GFP-tagged SUN1 variants (GFP-SUN1_785, GFP-SUN1_888, GFP-SUN1_916), or GFP alone. After incubation for 24 hours, total cell lysates were separated by a 10% acrylamide gel and analyzed by western blotting. (B) Cell migration activities were analyzed 24 hours after transfection. Transfection efficiency was ∼70% (determined using a Tali image cytometer) and all cells were counted including GFP-positive and -negative cells. Each bar represents the mean number of cells per field ±SD. *; p < 0.05, **; p < 0.01.
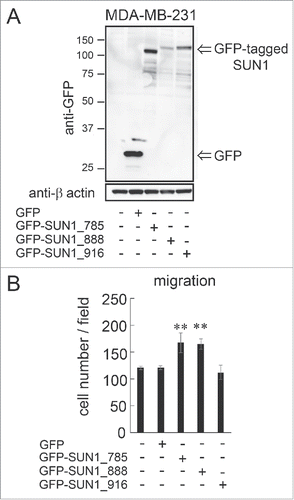
Figure 4. SUN1_888 but not SUN1_916 is required for cell migration. (A) Design of siRNA against SUN1 splice variants. (B) and (C). MDA-MB-231 cells were transfected with siRNA against SUN1_916, SUN1_888, total SUN1, or non-targeting siRNA and after 48 hours, total RNAs were analyzed by PCR (B) and migration activities were analyzed (C). Each bar represents the mean number of cells per field ±SD. *; p < 0.05, **; p < 0.01.
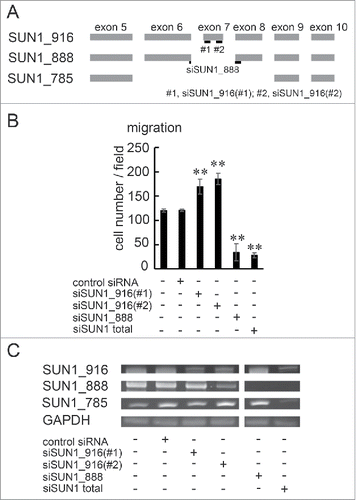
Figure 5. SUN1 interacts with B-type lamins. (A) Carboxy-terminus GFP-tagged SUN1 variants or GFP were transfected into HeLa cells and precipitated with anti-GFP pAbs. Then co-precipitated proteins were subjected to western blotting using indicated antibodies. (B) Carboxy-terminus GFP-tagged SUN1 variants or GFP-tagged SUN2 was transfected into HeLa cells and precipitated with anti-GFP pAbs. Then co-precipitated proteins were subjected to western blotting using anti-GFP, -lamin B1, and -lamin B2 antibodies. (C) The location of primer sets are indicated. Detailed sequence information is shown in Table S1. (D and E) Genomic DNA was obtained from parental HeLa, clone #1, or clone #2 cells. Then PCR was performed using indicated primer sets. Clone #1 showed single allele disruption, while clone #2 showed biallelic disruption. (F) SUN1 mRNA expression in parental HeLa, clone #1, and clone #2 cells was measured by real-time PCR. Results are presented as means ±SD. The values were normalized against GAPDH mRNA. (G) The expression level of SUN1 was analyzed by western blotting using anti-SUN1 pAbs and anti-actin mAb. (H) GFP tagged SUN1 variants or GFP were transfected into SUN1 knockout HeLa clone #2 cells and immunoprecipitation was performed with anti-GFP pAbs. Then precipitated proteins were subjected to western blotting using indicated antibodies.
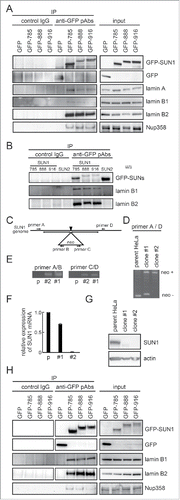
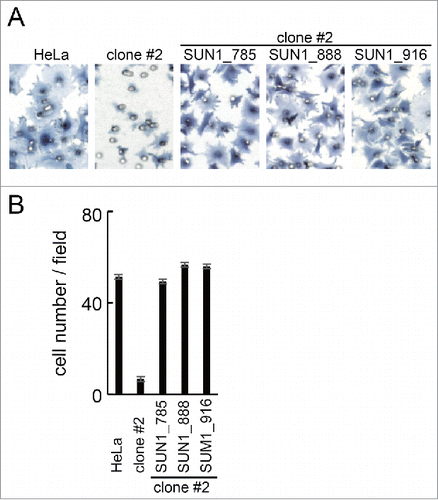
Figure 6. Rescued the cell migration of SUN1 knockout cells. (A) SUN1 knock out HeLa cells (clone #2) were transfected with GFP tagged SUN1 variants. Twenty-four hours after transfection, cell migration activities of parental HeLa cells, untransfected clone #2 and the SUN1 variants transfected cells were analyzed. Cells were stained with hematoxylin. Representative images are shown. Small holes in the images are pore of the membrane. (B) The number of migrated cells was counted. Each bar represents the mean number of cells per field ±SD.