Figures & data
Figure 1. Model of HLB assembly and function. HLBs are primarily organized by NPAT/Mxc and FLASH, which bind to the C terminus of NPAT/Mxc. HLBs in S phase (left) are larger than HLBs in other phases of the cell cycle (right), likely because Cyclin E/Cdk2 phosphorylation of NPAT/Mxc induces HLB reorganization and/or because mRNA synthesis via transcription and pre-mRNA processing is occurring within the HLB. Whereas some RD histone pre-mRNA processing factors like U7 snRNP and FLASH are constitutive residents of the HLB, other critical factors like the HCC are recruited only when histone genes are active. The assembly of the active cleavage complex (left) may utilize a pool of FLASH (and perhaps other factors) that is distinct from the pool of FLASH that binds to NPAT/Mxc and organizes the HLB. RNA pol II is enriched in the HLB, including in HLBs that are not actively synthesizing RD histone mRNA (J. Kemp and R. Duronio, unpublished). Whether concentrating RNA pol II in the HLB is functionally important and whether all the RNA pol II in the HLB is engaged in transcription during S phase are interesting open questions. Negative regulators of RD histone transcription like Drosophila Mute (GON4L/YARP in humans) are found concentrated only in the HLB, and likely modulate histone gene expression during the cell cycle in coordination with Cyclin E/Cdk2 activity. Note that the gene cluster at the top of the diagram is based on the arrangement of RD histone genes in Drosophila melanogaster, but conceptually applies to other RD histone gene clusters, which associate together in 3D space within the nucleus. Image created with BioRender.com.
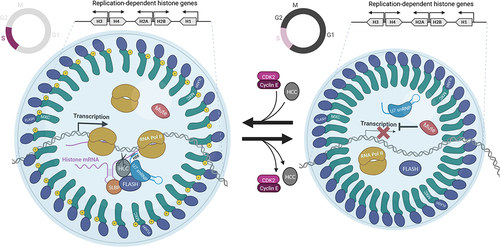
Figure 2. Active Drosophila HLBs display a core shell arrangement and are enriched in RNA polymerase II. High-resolution confocal images of Drosophila neuronal cells in the embryonic ventral nerve cord stained for Mxc (cyan), histone mRNA (magenta), and RNA polymerase II (yellow). The arrows indicate the HLB of two neuroblast stem cells in S phase as indicated by high level of cytoplasm histone mRNA (asterisks). Note the focus of nascent histone mRNA coincident with high amounts of RNA pol II that are both surrounded by a shell of Mxc protein. The arrowhead indicates an HLB in a quiescent cell that displays a more closed configuration of Mxc. Note that this HLB has RNA pol II, even though it is not in S phase, as indicated by the lack of RD histone mRNA. Scale bar = 2 microns.