Figures & data
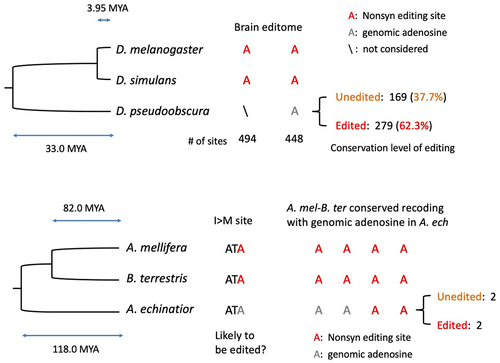
Figure 1. A-to-I RNA editing and the evolution of Adar auto-recoding sites in insects. (a) A-to-I RNA editing mediated by ADAR. I is recognized as G. (b) Prediction of adaptive RNA editing theory. The relative fitness of A- and G-alleles under different conditions is considered. (c) Drosophila Adar has an auto-recoding site that forms a negative feedback loop. (d) Evolution of Adar S>G (site1) and I>M (site2) sites in insects. “edSer”: editable serine codon; “unSer”: uneditable serine codon; “edIle”: editable isoleucine codon; “unIle”: uneditable isoleucine codon.
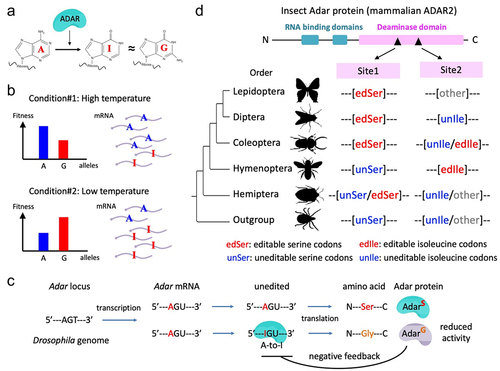
Figure 2. Observed and expected numbers/proportions of AA changes caused by nonsynonymous RNA editing. Total numbers of expected mutations were obtained by changing all adenosines to guanosines in the reference genome of D. melanogaster. The expected proportions of each AA change were estimated by the proportions of observed AA changes caused by nonsynonymous RNA editing. The 3-mer motif preferred by Adar is also considered. p values of the enrichments were calculated using one-tailed Fisher’s exact tests for each AA change. *, p < 0.05; ***, p < 0.001.
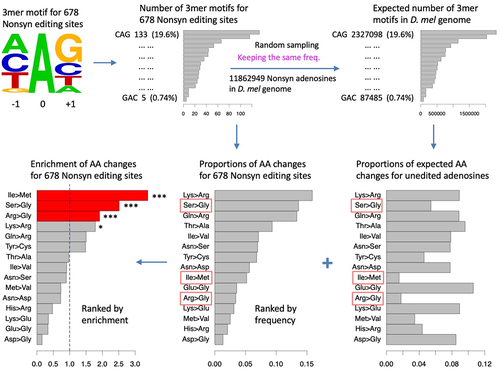
Figure 3. Enrichment of each type of AA change. (a) SNPs in global population of D. melanogaster. (b) Nonsynonymous editing sites in humans. (c) SNPs in global human populations.
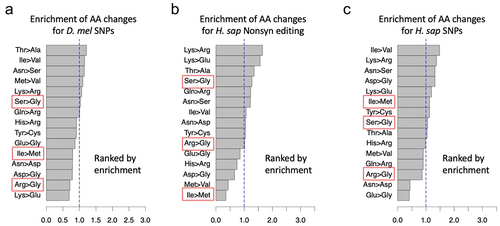
Figure 4. Hypotheses raised to explain the enrichment of Ile>Met, Ser>Gly, Arg>Gly recoding sites in Drosophila RNA editome. (a) the “uneditable-to-editable is beneficial” hypothesis. Editable is fitter than uneditable, but the benefit can either be diversifying or restorative. (b) The “deleterious editing replaced with uneditable codons” hypothesis. Since the editable ile, ser, and arg codons have another choice to mutate to uneditable ones, the observed editing after purifying selection shows enrichment on those codons.
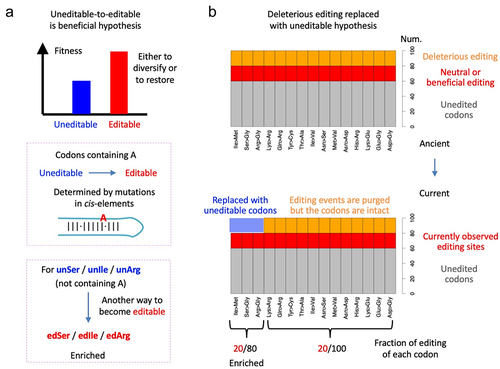
Table 1. Detailed information on the sequences used in this study.
Figure 5. The orthologous codons of Adar S>G (site1) and I>M (site2) auto-recoding sites in metazoan species with systematic RNA editing study. “#” means that although the codon itself is editable, no RNA editing events were reported by the original study. The phylogenetic tree is unscaled.
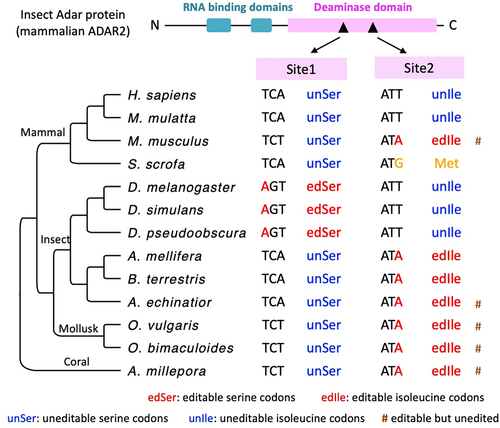
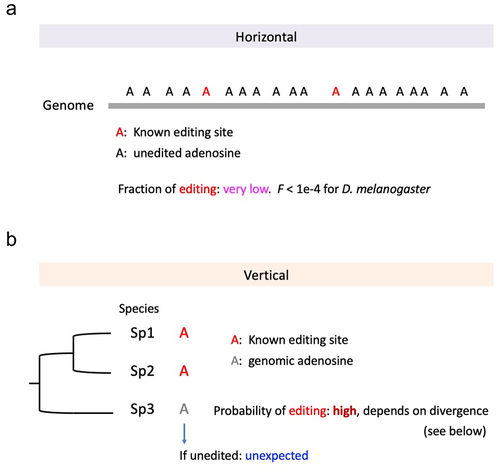
Figure 6. Horizontal and vertical comparison of editing sites. (a) the fraction of edited adenosines was very low in D. melanogaster genome. (b) Orthologous adenosine sites in three species. Two sites are known editing sites. The probability of the third species being edited should be high but the exact probability depends on the divergence.
Data availability statement
The analysis of the Adar auto-recoding codon involves the sequences in 13 species. Note that the Adar gene in insects is homologous to mammalian ADAD2 [Citation30]. In total, 13 representative species with systematic RNA editing studies were listed as follows (), including 6 from Arthropoda, 4 from Chordata, 2 from Mollusca, and Acropora millepora from Cnidaria as an outgroup. The nucleotide and protein sequences were downloaded from NCBI https://www.ncbi.nlm.nih.gov/ (accession IDs listed in ). The coding region of each mRNA sequence was extracted based on the annotation in NCBI.