Figures & data
Figure 1. NDGA blocked LPS-induced proinflammatory gene expression. (A–C) The expression of each gene in RAW264.7 cells was accessed using real-time PCR and the specific primers. The cells were pretreated with 10 µM NDGA for 24 hours, then challenged by LPS and cultured for 6 hour before the cell harvest. (D) The cells were transfected with pNF-κB-LUC and pRL-LUC, then 16 hours after transfection the cells were treated with 10 µM NDGA for 1 hour and subsequently stimulated with 100 ng/ml LPS for 3 hours. Firefly luciferase was normalized for Renilla luciferase activity.
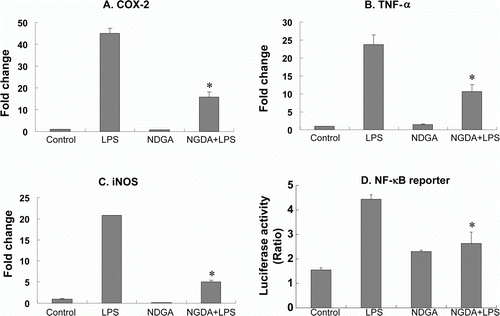
Figure 2. NDGA down-regulates TAK1 protein level. (A) Dose-dependent decreases in TAK1 level following the challenge of NDGA. RAW264.7 cells were treated with the indicated concentration of NDGA for 6 hours, then harvested and subjected to the western blot analysis. (B) The decreases in TAK1 level were shown in various cell lines. Each cell line was treated with 10 µM NDGA for 6 hours and analyzed for TAK1 level.
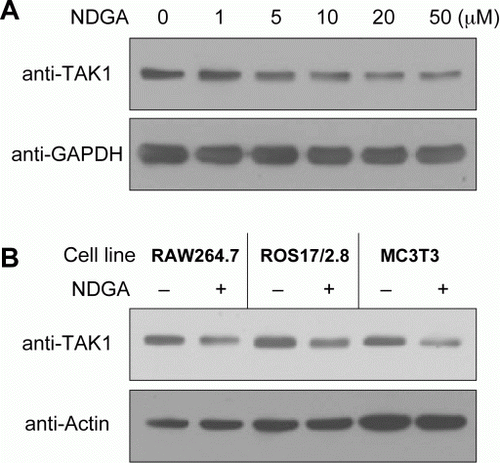
Figure 3. Effect of NDGA on TAK1 mRNA level. NDGA had no negative effect on TAK1 mRNA levels. RAW264.7 cells were treated with 10 µM NDGA for indicated times and were analyzed the expression of TAK1 mRNA by real-time PCR. Each mRNA level was expressed by comparing with 1-hour control level.
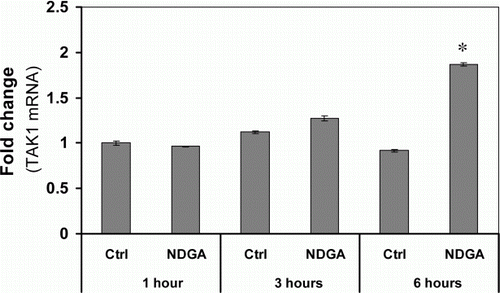
Figure 4. The involvement of proteasome in the reduction of TAK1 protein level caused by NDGA treatment. RAW264.7 cells were treated with 10 µM NDGA for indicated times in the absence (A) or in the presence (B) of 500 nM MG132.
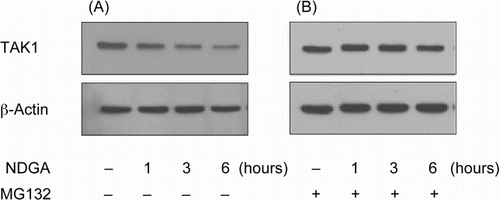
Figure 5. The time-course of NDGA effect on TAK1 protein level and on the related signaling pathway. The changes in the degradation of IκBα and phosphorylation of JNK upon LPS treatment with the variation of NDGA pretreatment time in RAW264.7 cell. NDGA was pretreated for various times, followed by LPS treatment. The cells were treated with 100ng/ml LPS for 1 hour and harvested for the western blot analysis.
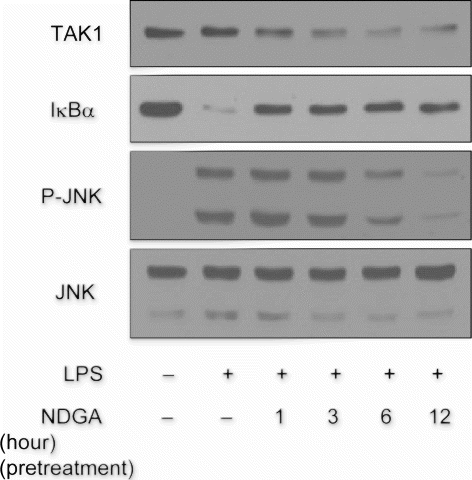
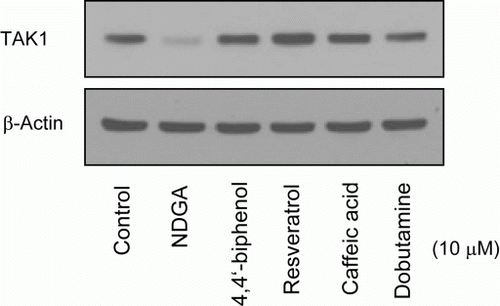
Figure 6. Effect of NDGA and its structurally related chemicals on the TAK1 protein level. RAW264.7 cells were treated with each compound (see for the molecular structure) at 10 µM concentration for 6 hours and subjected to the western blot analysis.