Figures & data
Figure 1. Functional domains and mutants of chromosome maintenance1 (CRM1). (A) Schematic structure of CRM1. Boxes 1–19 represent the HEAT repeat motifs, as defined by homology modeling. The CRIME domain (CRM1, importin β, etc.), which shares homology with importin β, and the acidic loop are involved in RanGTP binding. RanBP3-binding domain is indicated by green color. Modification of Cys528 by LMB targets the region involved in NES binding (Hamamoto et al. 1985; Nishi et al. 1994; Kudo et al. 1999). The C-terminal fragment corresponding to residues 707–1034 (CTR) is indicated by blue color. The consensus motif of Protein Kinase A (PKA) phpsphorylation site (Ser 1055) in the C-terminal domain is indicated above. The C-terminal GST fusion protein fragment region (960–1120 aa) is shown below. (B and C) Reciprocal Immunoprecipitation (IP) and Immunoblotting (IB) from HEK 293 cells. CRM1 and 14-3-3 immunoprecipitates were analyzed using anti-14-3-3 or anti-CRM1 antibody. Negative IP control was normal rabbit antibody. (D) Confocal microscopy. Endogenous CRM1 (red) or 14-3-3 (green) in HEK 293 cells was visualized using appropriate primary antibodies, and Alexa Fluor 568-conjugated secondary antibodies. Merged image (yellow) shows coincident distribution of wild type CRM1 and 14-3-3 proteins. Figures represent three or more independent experiments.
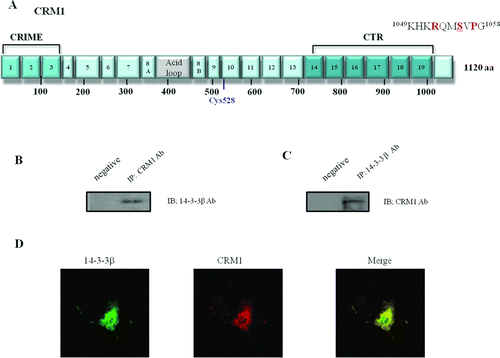
Figure 2. Formation of CRM1 and 14-3-3 protein–protein complexes. CRM1 and 14-3-3 immunoprecipitates were analyzed using anti-14-3-3 and anti-CRM1. CRM1 precipitated from HEK 293 cells (A). GST-tagged CRM1 mutants were expressed in E. coli and prebound to GST-agarose beads that were incubated with HEK 293 cell lysates and analyzed using anti-14-3-3 (B).
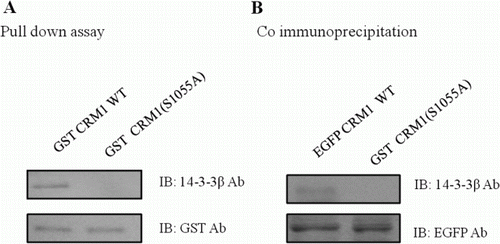
Figure 3. Phosphorylation of Ser 1055 in CRM1 by PKA was required for 14-3-3 binding. Plasmids with EGFP-CRM1 (WT) and EGFP-CRM1 S1055A were transfected into HEK 293 cells and CRM1 protein was immunopurified with GFP antibody. (A) Western blot with CRM1, 14-3-3, or phospho-Thr/Ser antibodies. CRM1 S1055A (lacking the PKA phosphorylation site) did not form a 14-3-3 protein complex or react with anti-phospho-Thr/Ser. (B) PKA assay with GST-CRM1 WT C-terminal fragment (amino acids 960–1120) or GST-CRM1 S1055A C-terminal fragment. GST-CRM1 WT, S1055A were expressed in E. coli and prebound to agarose beads that were incubated with PKA in assay buffer and analyzed using anti-phospho-Ser/Thr. Figures represent three independent experiments.
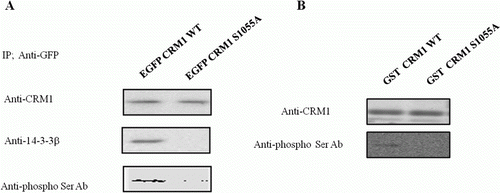
Figure 4. Sublocalization of CRM1 WT, S1055A, and S1055D with confocal microscopy. HEK 293 cells were transfected with plasmids for EGFP-CRM1, S1055A, or S1055D (green). Endogenous 14-3-3 proteins (red) were visualized using appropriate primary antibodies, and Alexa Fluor 568-conjugated secondary antibodies. Merged image (yellow) shows coincident distribution of wild type CRM1 and 14-3-3 proteins (A). No merged image (yellow) was seen with EGFP-CRM1 S1055A (B) or S1055D and 14-3-3 proteins (C). Figures represent three or more independent experiments.
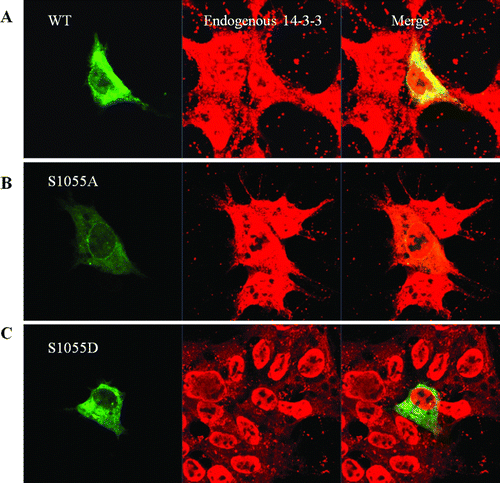
Figure 5. Subcellular localization of CRM1 phosphorylated by PKA. (A) HEK 293 cells were transfected with plasmids for EGFP-CRM1 WT, S1055A, or S1055D (green). CRM1 S1055D was detected in the cytoplasm as a large dot (right) and not on the nuclear rim. Figures represent three independent experiments. Confocal microscopic pictures were scanned using profile in the ZEN program. (B) Subcellular localization in HEK 293 cells of EGFP-CRM1 WT was examined after treatment with forskolin or H89 for 24 h. EGFP-CRM1 WT (green) was observed by fluorescence microscopy.
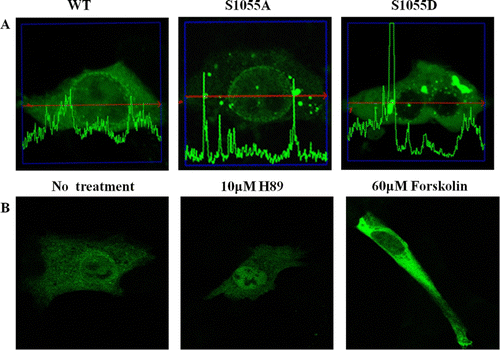
Table 1. Serine 1055 phosphorylation of CRM1 promoted cell apoptosis.
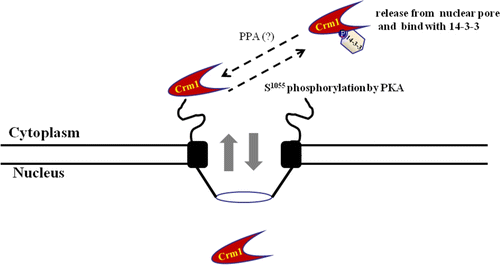
Figure 6. Schematic diagram of interaction between CRM1 and 14-3-3 proteins, with PKA. The nuclear pore export complex facilitates CRM1-dependent translocation of NES-containing proteins. Phosphorylation of CRM1 by PKA results in binding to 14-3-3 proteins as a new partner protein through the CRM1 C-terminal domain, which contains a conserved 14-3-3-binding motif (1049KHKRQMSVPG1058). Interaction regulates CRM1 nuclear pore localization and NES function in vivo and in vitro. Phosphorylated CRM1 seems to be dephosphorylated by a phosphoprotein phosphatase (PPA).