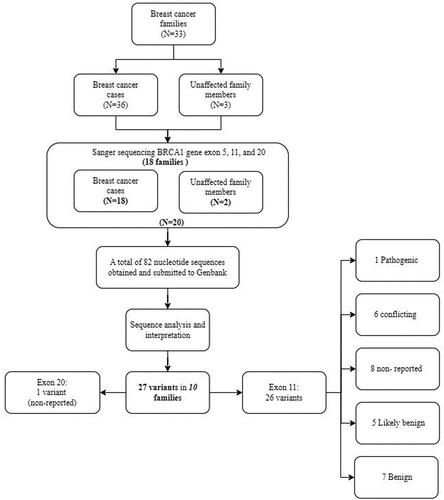
Figures & data
Table 1. Demographic and pathological features of the patients’ cohort.
Table 2. Pathogenicity status of BRCA1 germline variants identified in breast cancer families in this study and reporting status in ClinVar.
Table 3. Benign status of BRCA1 variants in this study.
Figure 2. Pedigrees of families with pathogenic, conflicting pathogenicity, likely benign, and non-reported clinically significant variants. A blackened circle represents an affected individual with breast cancer, while a divided circle represents an unaffected individual carrying variations. An affected individual with leukemia is represented by a green square, and one with colon cancer by an orange circle. The proband is indicated by an arrow.
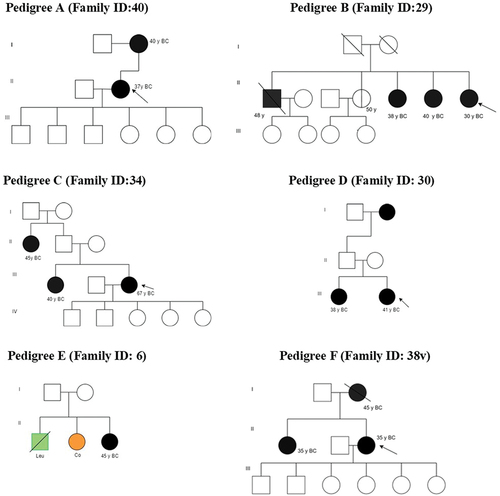
Table 4. Clinicopathological and molecular characteristics of BRCA1 variants.
Figure 3. Distribution and locations of identified BRCA1 variants. Only one variant was detected in exon 20. Exon 11 had 26 variants. One is pathogenic (c.2643del) and one is a novel allele (c.1366A>G). No variants were detected in exon 5. Vertical red indicate pathogenicity or conflicting interpretations of pathogenicity. Vertical green vertical lines indicate benign or likely benign variants.
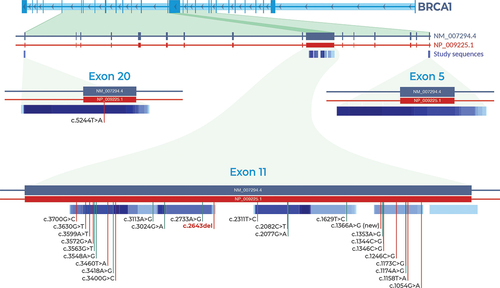
Table 5. Unclassified BRCA1 variants: frequency, predicted impact, and classification discrepancies.
supplemntary 3.xlsx
Download MS Excel (78.3 KB)supplementary1 .docx
Download MS Word (19.9 KB)Supplementary 2.docx
Download MS Word (16.5 KB)Data availability statement
All sequence data have been submitted to GenBank and provided with accession numbers (OR610584- OR610663) https://www.ncbi.nlm.nih.gov/nuccore/OR610584.