Figures & data
Table 1. Identification of periodontal pathogens in atherothrombotic samples using PCR methods
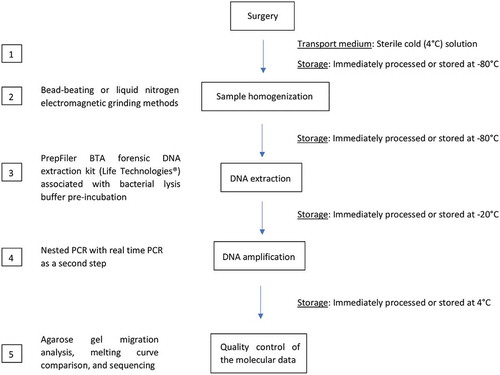
Table 2. Vascular sample preparation for identifying periodontal pathogens
Table 3. PCR conditions and identification methods for periodontal pathogens in atherothrombotic samples