Figures & data
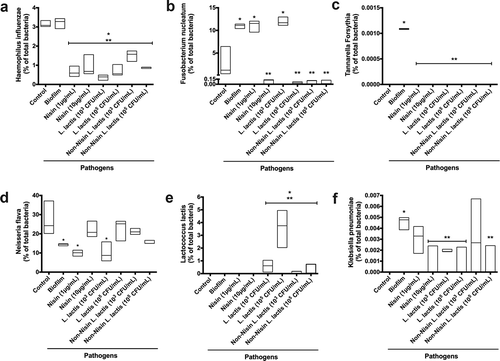
Figure 1. Nisin-producing probiotic inhibits oral biofilm formation, structure, and viability.
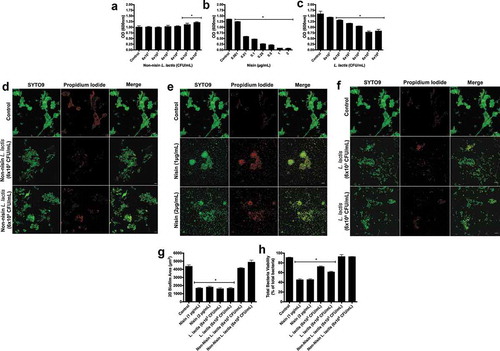
Figure 2. Nisin-producing probiotic disrupts oral biofilm formation, structure, and viability.
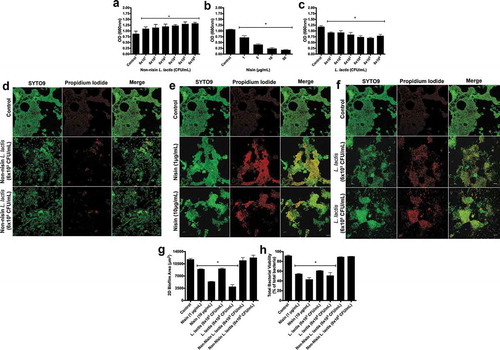
Figure 3. Nisin-producing probiotic disrupts pathogen-spiked oral biofilm formation, structure, and viability.
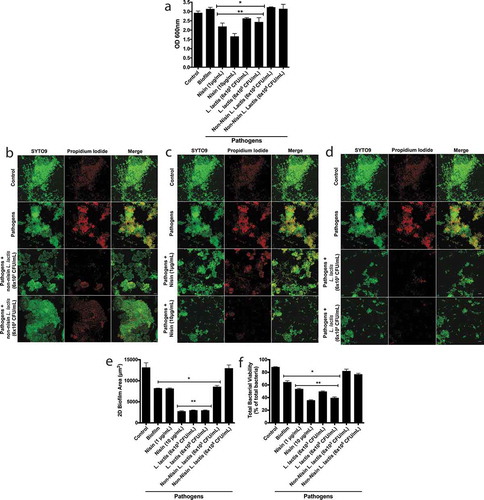
Figure 4. Biofilm bacterial diversity returns to control levels with nisin-producing probiotic or nisin.
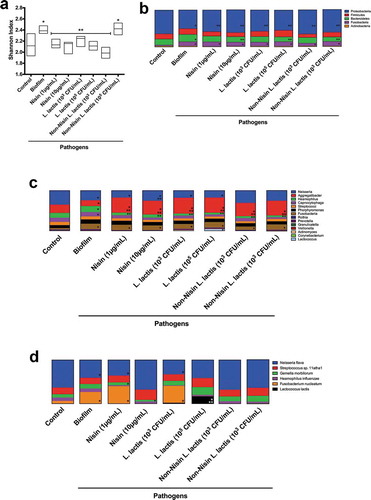
Figure 5. Nisin-producing probiotic and nisin significantly suppress F nucleatum, T. forsythia and K. pneumoniae, while promoting commensals in pathogen-spiked oral biofilms.