Figures & data
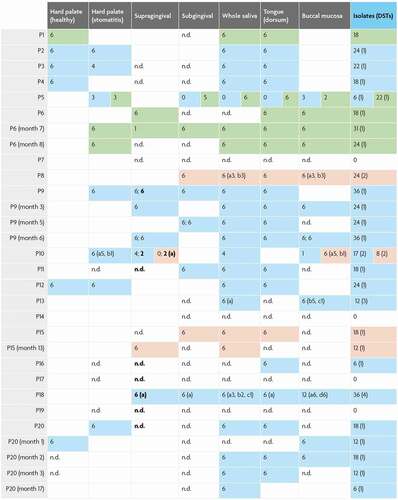
Table 1. C. albicans MLST DSTs for typed colonies. Diploid sequence types (DSTs) for C. albicans isolates showing the sequence type numbers for each sequenced region and clade membership and the number of colonies sequenced. DSTs have been submitted to the central MLST database; newly identified DSTs are highlighted green
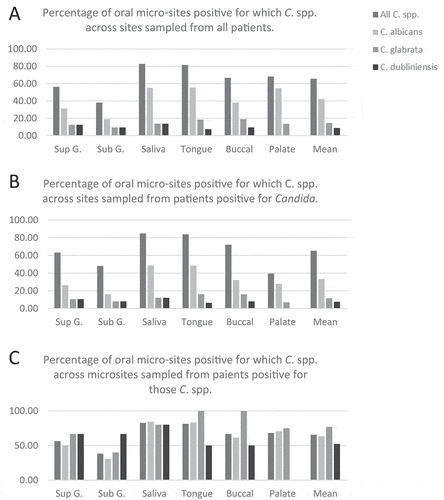
Figure 3. Population structure of C. albicans isolates. A) Phylogenetic network of C. albicans DSTs. Branches are coloured by clade as follows: 2, yellow; 12, green; 8, purple; 3, blue; 1, red; branches are grey where splits are shared. B) Cladogram (left) of P18 AAT1a microvariant sequences including the unphased ST (red) and both phased deconvolved sequences (blue) and nucleotide motif logos (right) for the unphased ST nucleotide variants (top) and phased haplotypes (bottom). C) eBURST analysis of C. albicans DSTs identified. The eBURST colouring convention for nodes is: founder, lime green; sub-founder, olive green; and for connecting line colours it is: high-level edges, gold; triple locus variant, red; double locus variant, green; branches are black and were without recourse to tiebreak rules
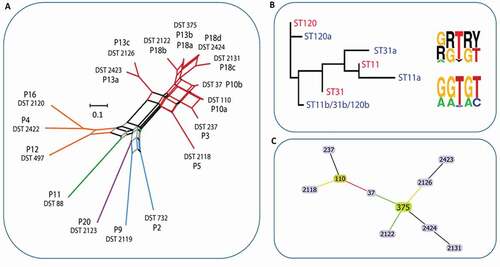
Figure 4. Population structure of C. dubliniensis and C. glabrata isolates. A) Phylogenetic tree of C. albicans and C. dubliniensis isolates identified including also representative strain types of the major C. albicans clades. The tree is produced from MLST sequences using the maximum likelihood method and is rooted at the black node, branches are gradient coloured bootstrap probabilities (red-high, blue-low), and nodes are sized by bootstrap support and coloured by branch length (red-short, blue-long). B) Neighbour joining phylogeny of the phylogenetic tree of C. glabrata isolates identified (grey bold type) including also all isolates present in the MLST database at the time of production (small black and coloured type). The tree is produced from MLST sequences using the neighbour joining method. STs representative of major groups are coloured: Group 1, pink; Group 2, blue; Group 3a, red; Group 3b, blue-grey; Group 5, green)
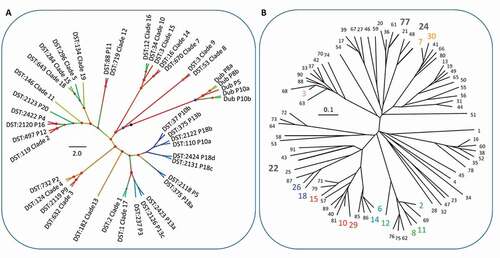
Table 2. C. glabrata MLST STs for typed colonies. Sequence types (STs) determined by MLST for C. glabrata isolates showing the sequence type numbers for each sequenced region and clade membership and the number of colonies sequenced. STs have been submitted to the central MLST database