Figures & data
Figure 1. RNA-seq results and RT-qPCR verification of EV-associated miRNAs.
(a) Volcano plot of relative representation of miRNAs in oligodendrocytes and oligodendrocyte-derived EVs. MiR-122-5p and miR-451a are strongly enriched in EVs (two top left dots marked), while miR-30d-5p and let-7g are equally represented in cells and EVs (n = 3). Red dotted line: significance level of 1% FDR (adjusted pValue = 0.01). logFC: log2 Fold Change, AveExpr: Average Expression. (b) RT-qPCR verification of miR-122-5p, miR-451a, miR-30d-5p and let-7g with independent biological replicates of cells and EVs (n = 3). Dashed line, cut-off Cq 35 defining background qPCR signal; error bars, SEM. (C) RNA-seq results indicate enrichment of the bovine- and primate-specific miR-1246 in EVs samples versus cells in all three replicates. RPM: Reads Per Million.
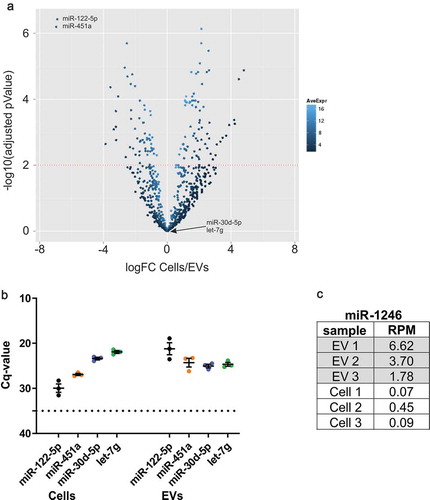
Figure 2. RT-qPCR analysis indicates miRNA contamination of defined media supplements.
(a) RT-qPCR of miRNAs derived from a pellet after differential ultracentrifugation (UC) of unconditioned basal medium (n = 3) or NS21 supplemented medium (n = 4, two different lots included, 2n each). Supplemented medium contains miR-122-5p, miR-451a, miR-30d-5p and let-7g, while basal medium is free of these miRNAs. Input volume for all samples: 25 ml. (b) RT-qPCR of miRNAs directly isolated from media supplements NS21 (n = 4, two different lots included, 2n each) and B27 (n = 2). Supplement input volume of 500 µl reflects amount represented in 25 ml used for UC-pelleted samples in (A). (c) RNase A treatment or combined Triton X-100/RNase A treatment of NS21 indicates partial protection of miR-122-5p to both treatment conditions. (d) RT-qPCR of miRNAs derived from supernatant and pellet fraction after depletion of NS21 supplement by ultracentrifugation. All miRNAs are detected in the pellet and supernatant indicating that depletion does not efficiently remove miRNAs from the supplement (n = 1). Dashed line, cut-off Cq 35 defining background qPCR signal; error bars, SEM.
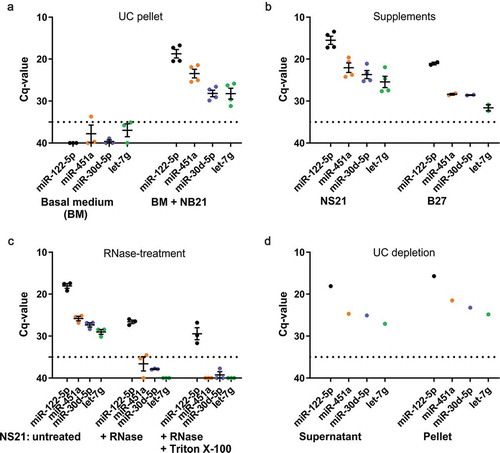
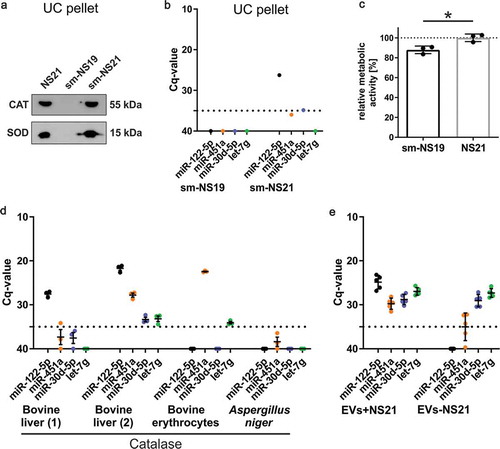
Figure 3. Analysis of NS21 supplement components reveals Catalase as the main source of miRNA contamination.
(a) Western blot analysis of pellets after application of differential ultracentrifugation (EV-isolation protocol) to non-conditioned media supplemented with commercial NS21, sm-NS19 lacking SOD and Catalase, or sm-NS21 including SOD and Catalase. Both SOD and Catalase (CAT) are detected in the pellet. (b) RT-qPCR of miRNAs derived from a pellet after differential ultracentrifugation of non-conditioned sm-NS19 lacking SOD and Catalase and sm-NS21 including SOD and Catalase. Sm-NS21 is positive for miR-122-5p indicating that SOD or Catalase addition is linked to miRNA contamination (n = 1). (c) MTT-assay to measure metabolic activity of primary oligodendrocytes cultured in sm-NS19 and sm-NS21. Lack of SOD and Catalase is associated with significantly reduced metabolic activity of the cells (n = 3, SEM, *p = 0.017, two-way ANOVA). (d) RT-qPCR analysis of miRNAs associated with Catalase. Mammalian Catalase derived from different suppliers and tissue origin is contaminated to different degrees with miRNAs, while Catalase from Aspergillus niger is miRNA-free (n = 3). (e) Differential RT-qPCR analysis of miRNAs associated with EVs collected in the presence (EV+NS21) or absence (EV-NS21) of NS21 supplement. miRNA-122-5p and −451a do only appear in EVs+NS21, while miR-30d-5p and let-7g are also present in EVs-NS21.