Figures & data
Figure 1. Study design.
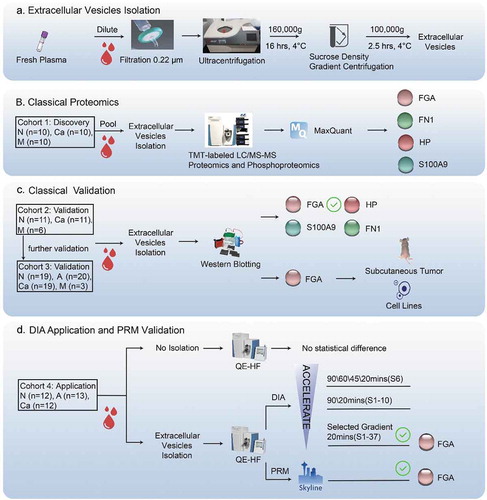
Figure 2. Characterization of plasma-derived EVs enriched from healthy donors and patients with CRC.
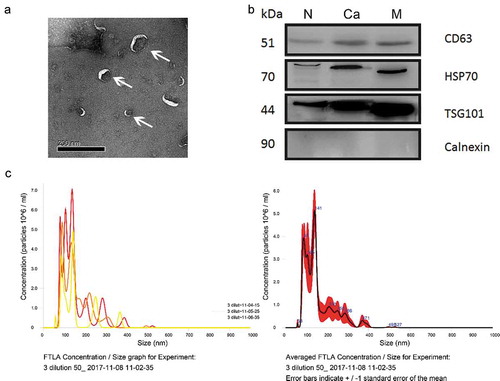
Figure 3. Proteomics and phosphoproteomics analyses of EVs.
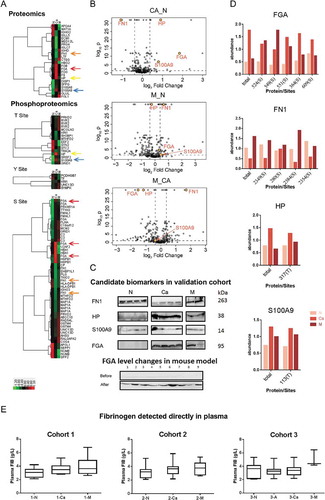
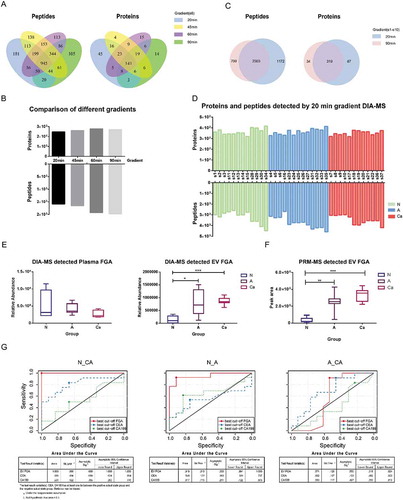
Figure 4. Clinical validation of DIA-MS and PRM-MS.