Figures & data
Table 1. Summary and comments of the result from the analysis of remains from 10 missing persons.
Figure 1. Mean and standard deviation of the coverage for each of the single nucleotide polymorphism (SNP) marker included in the analysis using massive parallel sequencing (MPS). The chart is based on the eight postmortem (PM) samples that resulted in full or partial SNP profiles.
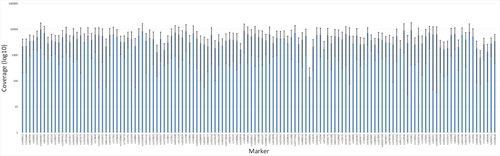
Figure 2. Sample and marker specific coverage for the eight postmortem (PM) samples that resulted in full or partial single nucleotide polymorphism (SNP) profiles for the massive parallel sequencing (MPS) analysis. A coverage of 200× was used as a minimum threshold for genotype calling.
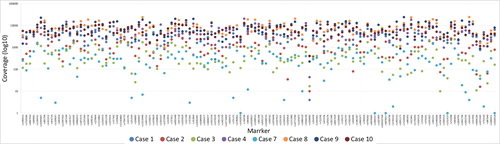
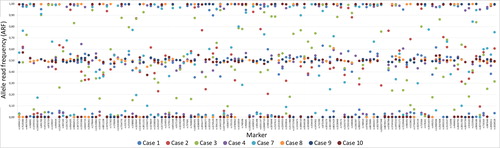
Figure 3. Sample and marker specific allele read frequency (ARF) for the eight postmortem (PM) samples that resulted in full or partial single nucleotide polymorphism (SNP) profiles for the massive parallel sequencing (MPS) analysis. An ARF values between 0.4 and 0.6 resulted in a heterozygous genotype and an ARF value between 0 and 0.1 or between 0.9 and 1 resulted in a homozygous genotype.