Figures & data
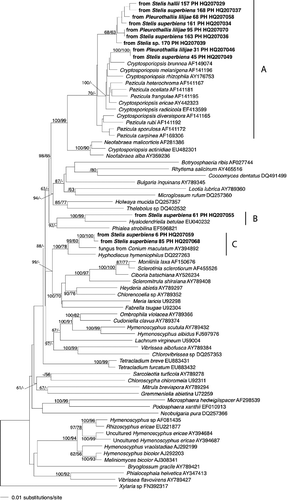
Figure 1. (A and B) Phylogenetic placement of ascomycete sequences obtained from the velamen of Stelis hallii, Stelis superbiens, Stelis concinna, Pleurothallis lilijaeand Stelis sp. Tree inferred from nuclear rDNA coding for the 5'-terminal domain of the large ribosomal subunit (nucLSU) using a maximum likelihood analysis implemented in RaxML with the GTRMIX algorithm. Numbers on branches designate Bayesian MCMC estimates of posterior probabilities and ML values (only values exceeding 50% are shown). The tree was rooted with Saccharomyces cerevisiaeEU649673.

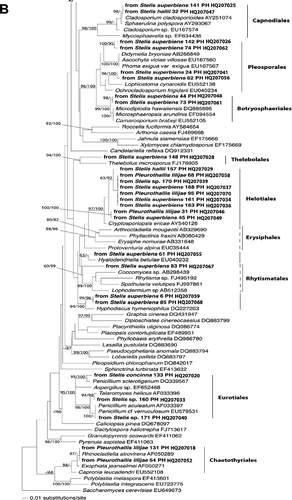
Table 1. Identified isolates from five epiphytic orchid species: Stelis hallii, S. superbiens, S. concinna, Stelis sp. and Pleurothallis lilijae. Numbers in parenthesis indicate the number of sampled individuals
Figure 2. Phylogenetic placement of Helotialean fungal isolate sequences from the velamen of Stelis hallii, Stelis superbiens, Stelis concinna, Pleurothallis lilijae and Stelis sp. Tree inferred from the ITS-5.8S region using a maximum likelihood analysis implemented in RaxML with the GTRMIX algorithm. Numbers on branches designate Bayesian MCMC estimates of posterior probabilities and ML values (only values exceeding 50% are shown). The tree was rooted with Xylaria sp. FN392317.