Figures & data
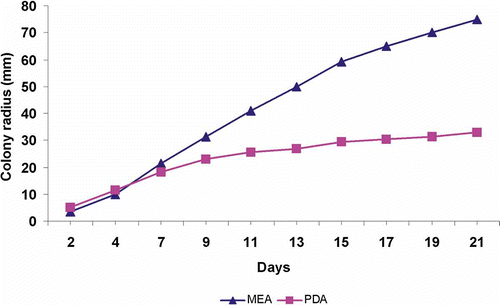
Figures 1–6. Neonothopanus nambi (BIN 2379). Figure 1. Basidiome in natural light and in darkness (copyright Dao Thi Van). Figure 2. Colonies on MEA and PDA media after 3 weeks growth. Figure 3. Colonies on MEA after 8 weeks growth. Figure 4. Broad hyphae and multiple clamps (DIC). Figure 5. Clamp connections and ring structure in narrow hyphae (DIC). Figure 6. Clamp connection (SEM).
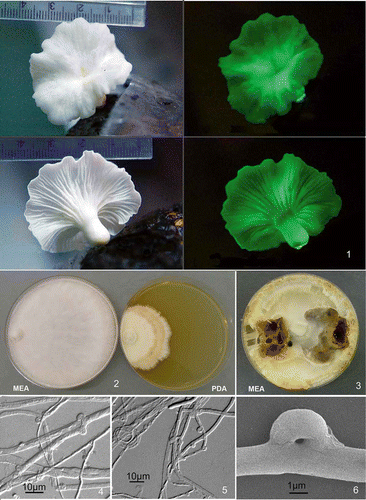
Figure 8. Parsimony analysis of Neonothopanus and clone sequences. Outgroup comprised of six Omphalotus taxa. Numbers above branches are bootstrap values >70%. The alignment comprised 42 taxa, length = 761 bp. A total of 169 characters were included in the analysis, including 110 parsimony informative.
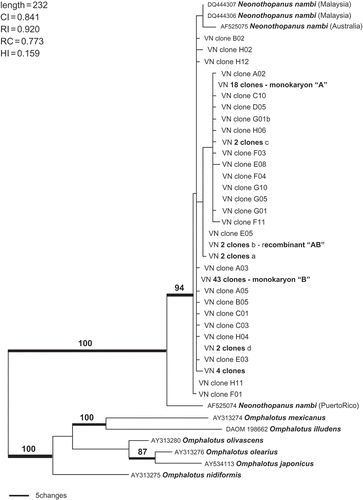
Figure 9. Alignment of Neonothopanus nambi clones. “Hallmarks” are nucleotide indels or single nucleotide polymorphisms inherited in the apparent recombinant, and also occurring in isolates from other geographic regions in varying recombinations. The numerical positions of the “hallmarks” in the alignment are indicted relative to the start of the ITS1. The location of multiple copy intragenomic nt variants are indicated by arrows with the copy numbers. “Singleton” clones are presumed cloning errors.
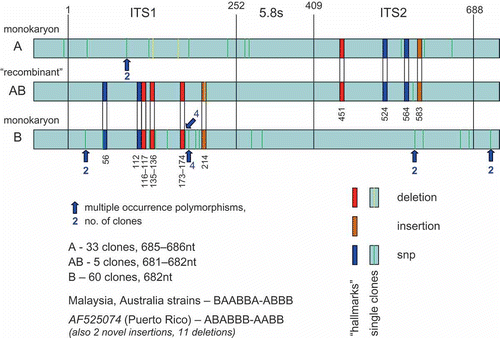
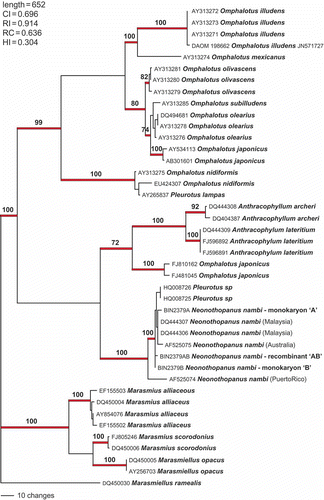
Figure 10. Phylogenetic analysis of available sequences for the Omphalotaceae. The outgroup comprises nine species of presumed Marasmiaceae. Numbers above branches are bootstrap values >70%. The alignment comprised 42 taxa; length 652 bp. A total of 359 characters were included in the analysis; 296 were parsimony informative.