Figures & data
Table 1. Yeast strains studied.
Figure 1. Direct sequencing chromatograms of the PCR product of the rDNA ITS1-5.8S-ITS2 region of Pichia membranifaciens strain CBS 215 using ITS1 (a) and ITS4 ((b), reverse complement of the original file) as the sequencing primers, showing the overlapped peaks.
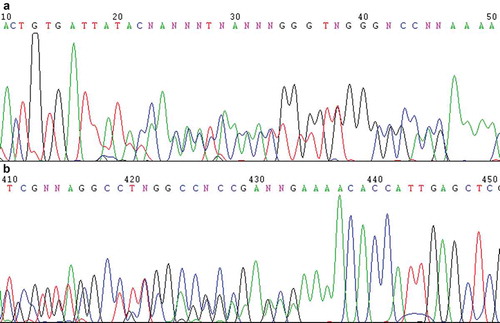
Figure 2. Sequence alignments of the different ITS types within the genome of Pichia membranifaciens strain CBS 215. At a given position, a nucleotide identical to that in the upper line is indicated by a dot, a gap is indicated by a hyphen.

Figure 3. Specific amplification of rDNA ITS types I–IV from the genomic DNA of strain CBS 215. M, molecular size mark.
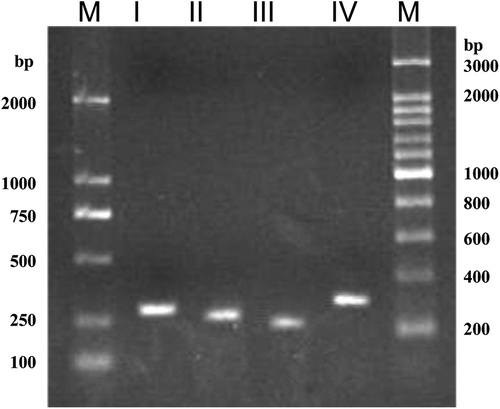
Table 2. Sequence mismatches of different ITS1 and 2 types of strain CBS 215 with the corresponding ITS regions of closely related strains of Pichia membranifaciens.
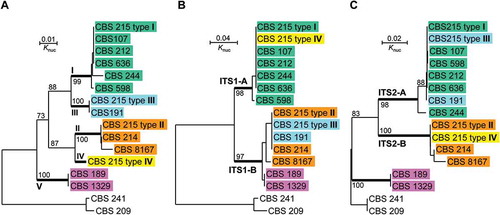
Figure 4. Neighbour-joining trees drawn from the sequence alignments of (a) the whole ITS repeat, (b) ITS1 only and (c) ITS2 only, of Pichia membranifaciens strains. Pichia manchurica strains CBS 209 and CBS 241 are used as the outgroup. Bootstrap percentages obtained from 1000 bootstrap replicates are shown.