Figures & data
Figure 1. The schematic representation showcases various approaches utilised in the study of chromatin modification. Techniques such as HPLC (High-Performance Liquid Chromatography), GC-MS (Gas Chromatography-Mass Spectrometry), and NMR (Nuclear Magnetic Resonance) spectroscopy are employed to gather information regarding alterations in metabolite profiling. These techniques contribute valuable insights into the characterisation and analysis of chromatin modifications.
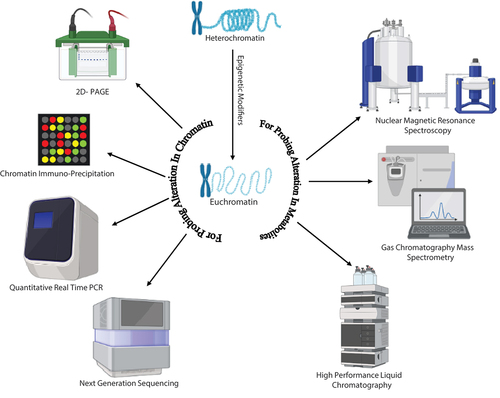
Figure 2. Chemical structure of epigenetic modifiers, such as 5-azacytidine (1), 5-aza-2’-deoxycytidine (2), suberohydroxamic acid (SBHA) (3), Sodium butyrate (4), Suberoylanilide hydroxamic acid (SAHA) (5), Trichostatin A (6), and Valproic acid (7) that are being used for the enhancement of secondary metabolite production. The chemical structure of metabolites obtained through epigenetic modifications, such as Cytosporone E (8), Dendrodolide E (9), Dendrodolide G (10), Dendrodolide I (11), Fumiquinazoline C (12), Vermistatin (13), Alternariol (14), Resveratrol (15), Eupenicinicol C (16), (Z)-9-octadecenoic acid (17), Dihydrovermistatin (18), 12-methyl-tetradecanoic acid (19), Cytosporone B (20), Isosulochrin (21), and Eupenicinicol D (22).
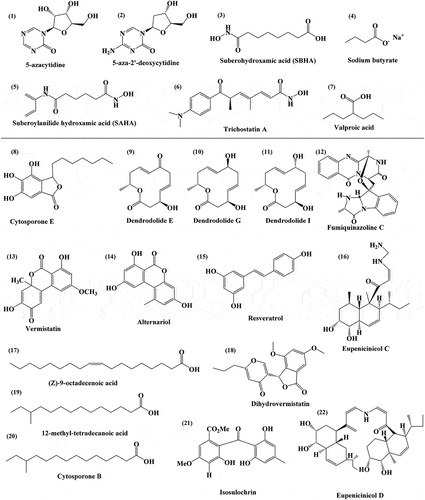
Table 1. List of epigenetic modifiers, their mode of action and elicited compounds.
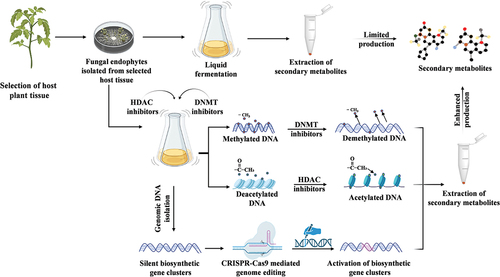
Figure 3. The diagram illustrates different strategies employed to enhance the production of fungal endophyte-associated metabolites. One approach involves the use of chemical elicitors to boost the production of secondary metabolites. Another approach utilises the CRISPR-Cas9 mediated genome editing technique, which enables targeted modifications to the fungal strain’s genome, ultimately leading to enhanced metabolite production.