Figures & data
Figure 1. A comparative structural analysis of the AF9-H3K9ac and Taf14-H3K9ac complexes. (A) An overlay of ribbon diagrams of the AF9 YEATS domain (cyan) in complex with the H3K9ac peptide (salmon) and the Taf14 YEATS domain (yellow) in complex with the H3K9ac peptide (gray). (B) The Taf14 (yellow) and AF9 (cyan) aromatic cages highlighting the interactions with K9ac. Red spheres represent water molecules, and apparent hydrogen bonds are indicated by blue dashes. For clarity backbone atoms were removed, unless involved in hydrogen bonds. (C) Superimposition of the H3K9ac peptides bound by Taf14 (gray) and AF9 (salmon) reveals that the RKac motif adopts a very similar conformation in both complexes. PDB IDs 4TMP and 5D7E.
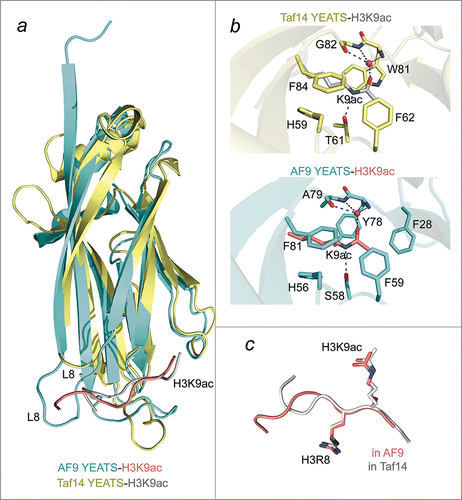
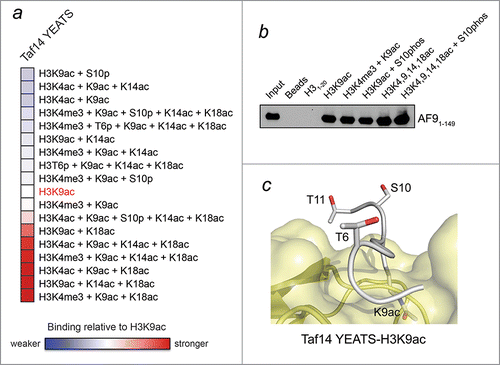
Figure 2. H3K9ac-YEATS interaction is modulated by neighboring histone PTMs. (A) Analysis of peptide microarray data from ref.Citation4 showing interactions between the YEATS domain of Taf14 and the indicated H3K9ac-containing peptides. Binding intensity is shown relative to H3K9ac without neighboring modifications (white). (B) Peptide pulldown and western blot analysis of the interaction between AF9-YEATS and the indicated peptides performed as previously described.Citation4 (C) The binding site of the Taf14 YEATS domain (yellow) for the H3K9ac peptide (gray).