Figures & data
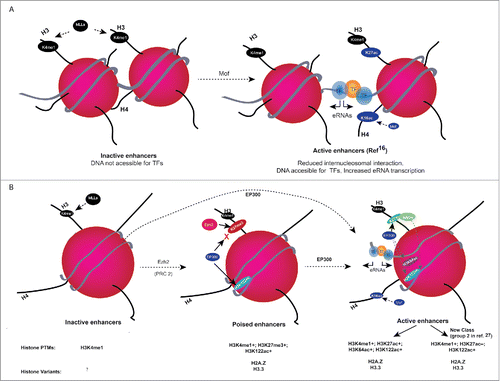
Figure 1. (A) H4K16ac role in increasing the accessibility of DNA to TFs basic amino acid residues at the histone H4 tail interaction with the acidic patches of H2A from neighboring nucleosomes, which is important for higher order chromatin folding. Mof-mediated acetylation of lysine 16 on the histone H4 (H4K16ac) tail disrupts the inter-nucleosomal interaction in vitro, which disrupts the higher order chromatin structure and might also increase the accessibility of TFs to DNA and increased eRNA transcription. (B) Working model for histone H3 globular acetylations at regulatory elements. Monomethylation of H3K4 residues (H3K4me1) away from transcription start sites (TSSs) is widely used to identify enhancers. Inactive enhancers are H3K4me1 positive, and they lack detectable level of histone acetylations. Poised enhancers are enriched for H3K27me3 along with H3K4me1 and H3K122ac. At active enhancers, EP300 acetylates nucleosomes at H3K27, H3K64, H3K122, and Mof acetylates H4K16. H3K122ac co-localizes with H2A.Z-containing nucleosomes, suggesting that H3K122ac might contribute to destabilization of nucleosomes at active and poised enhancers. Working model shows that EP300-mediated H3 globular domain acetylations lead to increased accessibility of nucleosomal DNA for TFs by reducing the affinity between histone and DNA. H3 globular acetylation facilitates the remodeling of nucleosomal DNA and contributes to destability of labile nucleosomes and eRNA transcription. Presence (+) or absence (−) of histone modifications, histone variants at enhancer classes is given below.