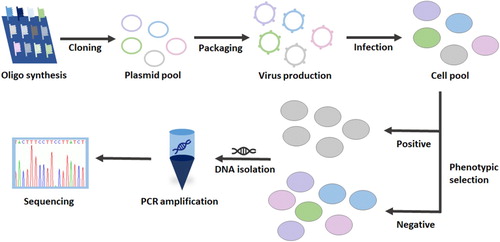
Figures & data
Figure 2. The concept of synthetic lethality In the simplest form, the simultaneous perturbation of two genes (shown here as gene A and B) leads to cell death, while perturbation of single gene does not. In the context of tumors, gene A can represent an oncogene and gene B can be identified as its synthetic lethal partner gene. The red crosses denote a mutation or a pharmacological inhibition.
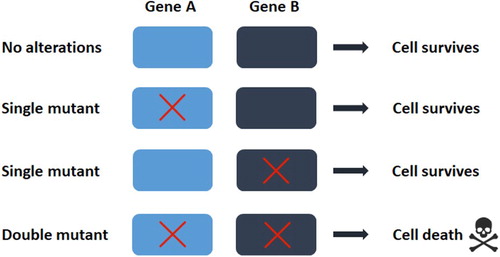
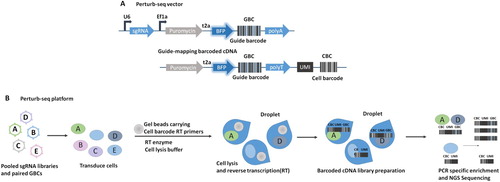
Figure 3. A robust strategy combining CRISPR screening with RNA-sequencing Schematic of Perturb-seq vector and guide-mapping barcoded cDNA. CBC (cell barcode) index for cell. UMI (unique molecular identifier), index for mRNA counting to correct for duplicates. GBC (guide barcode), index for sgRNA. (B) Schematic of the Perturb-seq platform.