Figures & data
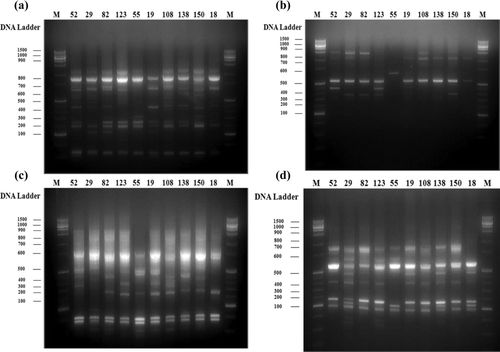
Table 1. Catechins content of selected tea trees used in this study.
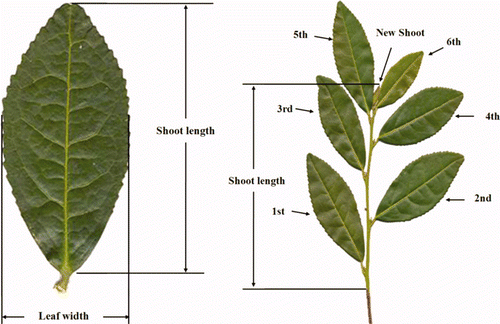
Table 2. Morphology analysis from selected tea tree lines.
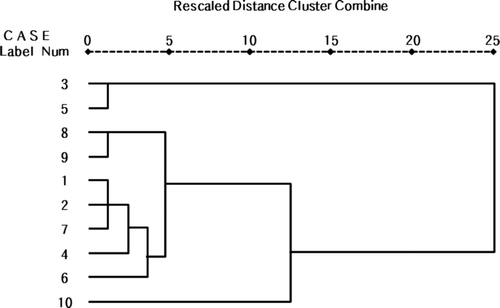
Figure 2. Morphology analysis from selected tea tree lines. (a) The average shoot length from selected tea tree lines. (b) The cluster analysis based on morphology analysis from selected tea tree lines. 1: HR-52, 2: HR-29, 3: HR-82, 4: HR-123, 5: HR-55, 6: HP-19, 7: HP-108, 8: HP-138, 9: HP-150 and 10: HP-18. This cluster analysis based on each variable was done by using the SPSS 17.0 statistics program.
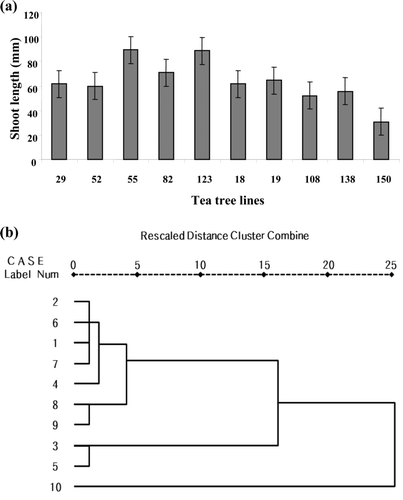
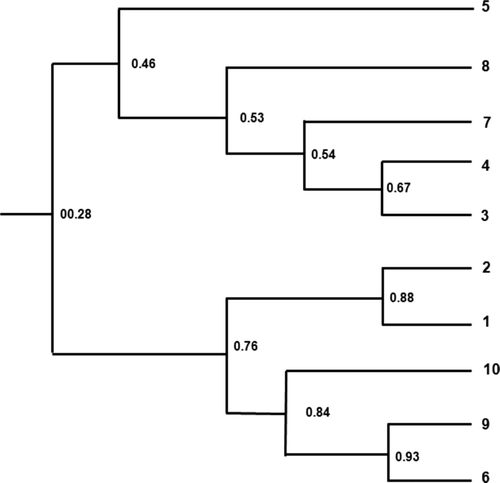
Figure 3. The cluster analysis based on four variables (leaf width, leaf length, leaf area, and shoot length) from selected tea tree lines. 1: HR-52, 2: HR-29, 3: HR-82, 4: HR-123, 5: HR-55, 6: HP-19, 7: HP-108, 8: HP-138, 9: HP-150, and 10: HP-18. This cluster analysis based on each variable was done by using the SPSS 17.0 statistics program.