Figures & data
Table 1. List of E. sylvestris samples used in this study.
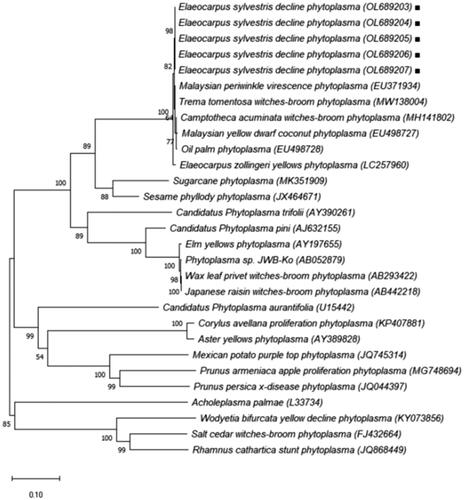
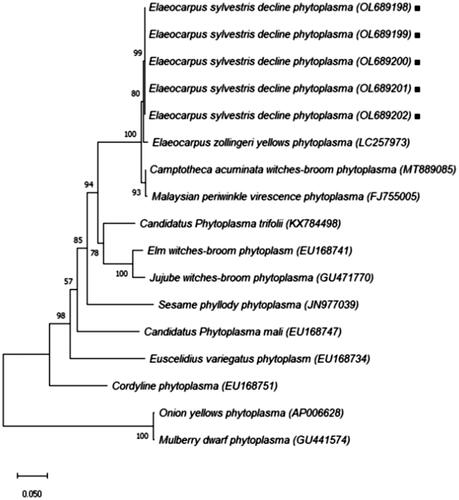
Table 2. Classification of phytoplasma species reference based on 16S rDNA gene.
Figure 1. Decline symptoms of Elaeocarpus sylvestris tree. (A) Yellowing leaves, (B) darkening leaves, (C) seeds near symptomatic leaves, (D) asymptomatic leaves, (E) decline symptoms tree in the park.

Figure 2. Agarose gel electrophoresis patterns of PCR products 16S rRNA gene and secA gene amplified by nested PCR using primer pair R16F2n/R16R2 and Direct SecAfor1/SecArev3 from E. sylvestris with decline symptoms in Jeju island, [Lane: M. molecular weight marker (100 bp), 1 ∼ 14: the symptomatic leaves and seeds, 15: jujube witches’ broom phytoplasma, 16: sumac witches’ broom phytoplasma].
![Figure 2. Agarose gel electrophoresis patterns of PCR products 16S rRNA gene and secA gene amplified by nested PCR using primer pair R16F2n/R16R2 and Direct SecAfor1/SecArev3 from E. sylvestris with decline symptoms in Jeju island, [Lane: M. molecular weight marker (100 bp), 1 ∼ 14: the symptomatic leaves and seeds, 15: jujube witches’ broom phytoplasma, 16: sumac witches’ broom phytoplasma].](/cms/asset/3d090da2-4c79-427f-bd33-2dc1ad154e0b/tfst_a_2029774_f0002_c.jpg)
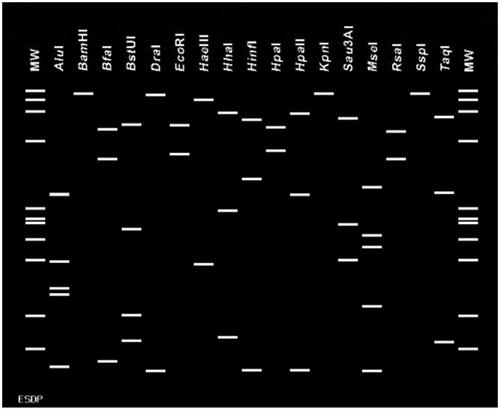
Figure 3. Virtual RFLP profiles of phytoplasma associated with E. sylvestris tree decline (Accession No: OL689203, OL689204, OL689205, OL689206 and OL689207) in-silico digestions. Sequences from strains ESDP-JJ1, JJ2, JJ3, JS1 and JS2 were used key restriction enzymes: AluI, BamHI, BfaI, BstUI, DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI, MseI, RsaI, SspI and TaqI were used to compare the RFLP pattern results. MW: ΦX174DNA digested with HaeIII.